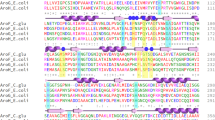
Abstract
The ArsA ATPase is the catalytic subunit of the ArsAB As(III) efflux pump. It receives trivalent As(III) from the intracellular metallochaperone ArsD. The interaction of ArsA and ArsD allows for resistance to As(III) at environmental concentrations. A quadruple mutant in the arsD gene encoding a K2A/K37A/K62A/K104A ArsD is unable to interact with ArsA. An error-prone mutagenesis approach was used to generate random mutations in the arsA gene that restored interaction with the quadruple arsD mutant in yeast two-hybrid assays. A number of arsA genes with multiple mutations were isolated. These were analyzed in more detail by separation into single arsA mutants. Three such mutants encoding Q56R, F120I and D137V ArsA were able to restore interaction with the quadruple ArsD mutant in yeast two-hybrid assays. Each of the three single ArsA mutants also interacted with wild type ArsD. Only the Q56R ArsA derivative exhibited significant metalloid-stimulated ATPase activity in vitro. Purified Q56R ArsA was stimulated by wild type ArsD and to a lesser degree by the quadruple ArsD derivative. The F120I and D137V ArsAs did not show metalloid-stimulated ATPase activity. Structural models generated by in silico docking suggest that an electrostatic interface favors reversible interaction between ArsA and ArsD. We predict that mutations in ArsA propagate changes in hydrogen bonding and salt bridges to the ArsA–ArsD interface that affect their interactions.




Similar content being viewed by others
References
Abernathy CO, Liu YP, Longfellow D, Aposhian HV, Beck B, Fowler B, Goyer R, Menzer R, Rossman T, Thompson C, Waalkes M (1999) Arsenic: health effects, mechanisms of actions, and research issues. Environ Health Perspect 107(7):593–597
Adams A, Gottschling DE, Kaiser C, Stearns T (1998) Methods in yeast genetics: a cold spring harbor laboratory course manual. Cold Spring Harbor laboratory, Cold Spring Harbor
Auld KL, Hitchcock AL, Doherty HK, Frietze S, Huang LS, Silver PA (2006) The conserved ATPase Get3/Arr4 modulates the activity of membrane-associated proteins in Saccharomyces cerevisiae. Genetics 174(1):215–227
Beane Freeman LE, Dennis LK, Lynch CF, Thorne PS, Just CL (2004) Toenail arsenic content and cutaneous melanoma in Iowa. Am J Epidemiol 160(7):679–687
Bhattacharjee H, Rosen BP (1996) Spatial proximity of Cys113, Cys172, and Cys422 in the metalloactivation domain of the ArsA ATPase. J Biol Chem 271(40):24465–24470
Bhattacharjee H, Li J, Ksenzenko MY, Rosen BP (1995) Role of cysteinyl residues in metalloactivation of the oxyanion-translocating ArsA ATPase. J Biol Chem 270(19):11245–11250
Bozkurt G, Stjepanovic G, Vilardi F, Amlacher S, Wild K, Bange G, Favaloro V, Rippe K, Hurt E, Dobberstein B, Sinning I (2009) Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc Natl Acad Sci USA 106(50):21131–21136
Cadwell RC, Joyce GF (1992) Randomization of genes by PCR mutagenesis. PCR Methods Appl 2(1):28–33
Chen CM, Misra TK, Silver S, Rosen BP (1986) Nucleotide sequence of the structural genes for an anion pump. The plasmid-encoded arsenical resistance operon. J Biol Chem 261(32):15030–15038
DeLano WL (2001) The PyMOL user’s manual. DeLano Scientific, San Carlos
Denic V, Dotsch V, Sinning I (2013) Endoplasmic reticulum targeting and insertion of tail-anchored membrane proteins by the GET pathway. Cold Spring Harb Perspect Biol 5(8):a013334
Fields S, Song O (1989) A novel genetic system to detect protein–protein interactions. Nature 340(6230):245–246
Gill SC, von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182(2):319–326
Green MR, Sambrook J, Sambrook J (2012) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Hirst M, Ho C, Sabourin L, Rudnicki M, Penn L, Sadowski I (2001) A two-hybrid system for transactivator bait proteins. Proc Natl Acad Sci USA 98(15):8726–8731
Hsu CM, Rosen BP (1989) Characterization of the catalytic subunit of an anion pump. J Biol Chem 264(29):17349–17354
Hua SB, Qiu M, Chan E, Zhu L, Luo Y (1997) Minimum length of sequence homology required for in vivo cloning by homologous recombination in yeast. Plasmid 38(2):91–96
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):27–38
Joshi PB, Hirst M, Malcolm T, Parent J, Mitchell D, Lund K, Sadowski I (2007) Identification of protein interaction antagonists using the repressed transactivator two-hybrid system. Biotechniques 42(5):635–644
Klopotowski T, Wiater A (1965) Synergism of aminotriazole and phosphate on the inhibition of yeast imidazole glycerol phosphate dehydratase. Arch Biochem Biophys 112(3):562–566
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372(3):774–797
Lin YF, Walmsley AR, Rosen BP (2006) An arsenic metallochaperone for an arsenic detoxification pump. Proc Natl Acad Sci USA 103(42):15617–15622
Lin YF, Yang J, Rosen BP (2007) ArsD residues Cys12, Cys13 and Cys18 form an As(III) binding site required for arsenic metallochaperone activity. J Biol Chem 282(23):16783–16791
Macindoe G, Mavridis L, Venkatraman V, Devignes MD, Ritchie DW (2010) HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res 38(Web Server issue):W445–449
Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, Hegde RS, Keenan RJ (2009) The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461(7262):361–366
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26(16):1781–1802
Rosen BP, Weigel U, Karkaria C, Gangola P (1988) Molecular characterization of an anion pump. The arsA gene product is an arsenite (antimonate)-stimulated ATPase. J Biol Chem 263(7):3067–3070
Ruan X, Bhattacharjee H, Rosen BP (2006) Cys-113 and Cys-422 form a high affinity metalloid binding site in the ArsA ATPase. J Biol Chem 281(15):9925–9934
Ruan X, Bhattacharjee H, Rosen BP (2008) Characterization of the metalloactivation domain of an arsenite/antimonite resistance pump. Mol Microbiol 67(2):392–402
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234(3):779–815
San Francisco MJ, Hope CL, Owolabi JB, Tisa LS, Rosen BP (1990) Identification of the metalloregulatory element of the plasmid-encoded arsenical resistance operon. Nucleic Acids Res 18(3):619–624
Shen J, Hsu CM, Kang BK, Rosen BP, Bhattacharjee H (2003) The Saccharomyces cerevisiae Arr4p is involved in metal and heat tolerance. Biometals 16(3):369–378
Tisa LS, Rosen BP (1990) Molecular characterization of an anion pump. The ArsB protein is the membrane anchor for the ArsA protein. J Biol Chem 265(1):190–194
Vogel G, Steinhart R (1976) ATPase of Escherichia coli: purification, dissociation, and reconstitution of the active complex from the isolated subunits. Biochemistry 15(1):208–216
Yang J, Rawat S, Stemmler TL, Rosen BP (2010) Arsenic binding and transfer by the ArsD As(III) metallochaperone. Biochemistry 49(17):3658–3666
Yang J, Abdul Salam AA, Rosen BP (2011) Genetic mapping of the interface between the ArsD metallochaperone and the ArsA ATPase. Mol Microbiol 79(4):872–881
Ye J, Ajees AA, Yang J, Rosen BP (2010) The 1.4 Å crystal structure of the ArsD arsenic metallochaperone provides insights into its interaction with the ArsA ATPase. Biochemistry 49(25):5206–5212
Zhou T, Rosen BP (1997) Tryptophan fluorescence reports nucleotide-induced conformational changes in a domain of the ArsA ATPase. J Biol Chem 272(32):19731–19737
Zhou T, Radaev S, Rosen BP, Gatti DL (2000) Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO J 19(17):4838–4845
Zhou T, Radaev S, Rosen BP, Gatti DL (2001) Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation. J Biol Chem 276(32):30414–30422
Zhu Y-G, Yoshinaga M, Zhao F-J, Rosen BP (2014) Earth abides arsenic biotransformations. Annu Rev Earth Planet Sci 42:443–467
Acknowledgments
This study was supported by National Institutes of Health Grant GM55425.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pillai, J.K., Venkadesh, S., Ajees, A.A. et al. Mutations in the ArsA ATPase that restore interaction with the ArsD metallochaperone. Biometals 27, 1263–1275 (2014). https://doi.org/10.1007/s10534-014-9788-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-014-9788-6