Abstract
Dynamics of dissolved organic matter (DOM) in ecosystems are controlled by a suite of interacting physical, chemical, and biological factors. Growing recognition of the associations between microbial communities and metabolism and intrinsic DOM characteristics, highlight the potential importance of microbe-DOM relationships to modulate the role and fate of DOM, yet these relationships are difficult to isolate because they often operate across confounding environmental gradients. In a controlled laboratory incubation (44 days), we integrated DOM bulk and molecular characterization, bacterial abundances, microbial assemblage composition, nutrient concentrations, and cellular respiration to discern the structural dynamics of biological processing among DOM sources from different allochthonous litters (grass, deciduous leaves, and evergreen needles). We identified two periods, consistent among DOM sources, where processing dynamics differed. Further, bulk fluorescent analyses showed shifts from low to high excitation and emission wavelengths, indicating the biological production of more complex/degraded materials over time. Molecular level analyses revealed similar temporal patterns among DOM sources in the production and consumption of individual chemical components varying in reactivity and heteroatomic content. Despite these similarities, total carbon (C) removed and carbon dioxide (CO2) accumulation differed by ~ 20% and 25% among DOM sources. This range in C processing was apparently tied to key chemical properties of the DOM (e.g., initial DOM composition, N content, and labile nature) as well as differential reorganization of the microbial populations that decomposed the DOM. We conclude that the production, transformation, and consumption of C in aquatic ecosystems is strongly dependent on the source and character of DOM as well as the structure of the microbial communities present, both of which change as DOM is processed over time. It is crucial that stream C processing models represent this complexity accurately.
Similar content being viewed by others
Introduction
Dissolved organic matter (DOM) represents the dominant flux of carbon (C) and nutrients in many aquatic ecosystems (Fisher and Likens 1973; Meyers and Lallier-Verges 1999). As such, DOM is crucial for a variety of ecological and physicochemical processes in aquatic ecosystems including heterotrophic production and the coupling of elemental cycles (Bernhardt and Likens 2002; Brookshire et al. 2005; Seidel et al. 2016; Vannote et al. 1980; Wetzel 1995). However, the degree to which DOM is biologically reactive, and subsequently processed, depends on a myriad of factors such as the initial DOM composition, physicochemical conditions of the environment, and the biological communities (Mineau et al. 2016). Given the growing appreciation of lotic ecosystems’ role in biogeochemical cycling (Cole et al. 2007; Hotchkiss et al. 2015), elucidating how the above factors influence DOM processing is necessary to determine when in-stream processing is influential and contributes to broad scale fluxes of energy and nutrients.
Elucidating the dynamics of DOM through time is inherently challenging because of the interwoven nature of the physical, chemical, and biological variables that determine its processing in the environment. Measuring bulk C concentrations over various temporal scales provides information about net material fluxes but fails to address what types of C are being cycled, when, and how. This is crucial to identify DOM utilization, production, and transformation mechanisms within and across ecosystems. Identifying and describing patterns of DOM composition, structure, character (e.g., amino acid- versus lignin-like, etc.), and reactivity (labile and recalcitrant) provides details into how DOM is created, conserved, and transformed (Fellman et al. 2010; Gonsior et al. 2009; Jaffé et al. 2008; Lam et al. 2007; Maie et al. 2005; Osburn et al. 2001). Specifically, the intrinsic molecular level DOM composition (CcHhNnOoSs containing compounds) and properties (e.g., the proportion of reactive constituents, heterogeneous nature, chemical structure, and molecular weights) set the boundaries for microbial community dynamics and transformations. Therefore, complementary analyses incorporating bulk and molecular level assessments of quantity and quality can reveal important connections among the DOM pool, its underlying chemistry, and biological processes.
Microbial community assemblages and DOM composition are inherently linked. DOM properties govern microbial community composition and metabolic processes that consume, transform, and produce different DOM composition, thereby altering properties of the DOM pool. There is growing recognition of the interconnected nature of DOM and microbes (Amaral et al. 2016; Fasching et al. 2014; Logue et al. 2016; Smith et al. 2018), however, we are still developing an understanding of how combining different qualitative and quantitative measures of DOM can be linked to microbial community structure to help predict response (e.g., heterotrophic respiration rates, DOM consumption, and carbon dioxide [CO2] accumulation). Pairing microbial data with qualitative DOM composition measurements in a temporally explicit framework allows us to begin to identify key factors in the microbial-DOM composition feedback loop. In this study, we let the microorganisms answer two questions, “what type of DOM is preferred?” and “when is it utilized?” This approach has the potential to reveal substantive insights into the dynamics of DOM transformation and removal.
Here, we couple biological and chemical techniques to relate biological processing with DOM composition through time. Specifically, our objective was to use a controlled, small-scale microcosm experiment to isolate temporal patterns of C processing among three allochthonous DOM sources. We were interested in the following overarching question: “How are differences in biological function (i.e. respiration and total C utilization) related to DOM composition through time?” To address this, we integrated direct measurements of biological structure and function with multiple DOM chemical characterization techniques at the bulk and molecular level over a 44-day period.
Materials and methods
Sample collection, processing, and experimental design
Plant litter and stream water were collected from Sourdough Creek, a third order stream located in southwestern Montana, USA (45°38′06.86 N, 111°01′54.28 W). At the collection site, the stream flows through areas of mixed land cover, including woodland, agriculture, and suburban. Stream dissolved organic carbon (DOC) concentration prior to incubation was 1.46 mg/L. Nutrient concentrations in the Sourdough Creek are low with concentrations of dissolved inorganic nitrogen (DIN) from below detection to 0.17 mg/L and phosphate concentrations ranging from 0.008 to 0.054 mg/L (detection limits 0.001–0.005 mg/L for nitrate, nitrite, and phosphorus; Montana Department of Environmental Quality: http://deq.mt.gov/).
In autumn of 2013, we collected newly abscised or senesced organic litter from three plant species prevalent along the Sourdough Creek riparian corridor, Trembling aspen (Populus tremuloides; “leaf”), Brome grass (Bromus sp.; “grass”), and Douglas-Fir (Pseudotsuga menziesii, “needle”). Plant litter was dried at room temperature (25 °C) and ground into coarse fragments. We created DOM extracts of each litter species by leaching approximately 10 g of litter fragments for 24 h in 2 L of deionized water at 29 °C, shaking every 6 h. After leaching, particulate organic matter and microbes were removed by filtering 1 L through pre-combusted 0.45 µm glass fiber filters. A set of three 1 L glass bottles were used for each litter treatment incubation. Varying volumes of DOM extracts (leaf: 42 mL, needle: 110 mL, and grass: 129 mL) were added to combusted 1 L glass bottles with 0.5 L Sourdough Creek stream water (including local microbial consortia), and the remaining volume filled with Milli-Q water to standardize C concentration for all samples to a target concentration of 20 mg/L. DOM extract accounted for ~ 95% of total DOC. Bottles were incubated at 29 °C in the dark over 44 days at which point, > 75% of the initial DOC had been consumed and consumption was < 0.25%/day. To isolate and quantify the biological processing of different C sources without confounding variables (e.g., changes in temperatures, pH, light exposure, oxygen conditions, and microbial consortia) (sensu Pastor et al. 2018; Vonk et al. 2015), the incubation bottles were not continuously aerated over time, creating conditions identical to the respirometry measurements. One bottle was sampled and resealed at each time point for bulk measurements. Sampling schemes for bacterial abundances, DOC and major ion concentrations, and optical spectroscopy measurements included days 0, 1, 2, 4, 16, 30, 37, and 44 (destructively sampled from one bottle). Molecular composition samples for Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS) analysis were collected for the stream water, initial leachate extracts, and on days 0, 2, and 44. Samples for next generation sequencing were collected on the stream water and at day 2 for each leachate C source. One bottle each for FT-ICR MS and next generation sequencing measurements were destructively sampled. The lack of replication for the incubations was a practical consideration based on the amount of sampling time points and response variables measured. Respirometry experiments (see below) were conducted in triplicate combusted glass bottles and sampled every 12 h over 44 days.
Bulk elemental concentration of particulate plant litter
Each litter type was measured for C, nitrogen (N), and phosphorus (P) content, which were used to calculate C:N, C:P, and N:P elemental stoichiometry. Plant litter was homogenized using a mortar and pestle and subsampled for analyses. All subsamples of C and N content were analyzed on a Costech Elemental Analyzer (Costech Inc., USA), with wheat as an external standard (average recovery was 98% and 97% for %C and %N, respectively). P content was quantified using persulfate digestion and the ascorbic acid method (APHA 1992) with bovine muscle as an external standard (average recovery 92%). Elemental ratios presented are on a molar basis.
Bulk C and major ion concentrations
DOC concentration was measured after filtration through a 0.2 μm low-carbon filter into a clean, combusted amber glass vial by a GE Sievers 900 Portable Total Organic Carbon Analyzer. Samples were analyzed immediately after collection. DOC concentrations were measured with analytical triplicates (sample size 25 mL), and Milli-Q water blanks were run between each sample to prevent cross-contamination. Leachate extracts, experimental samples, and blanks were measured four times with one analytical reject prior to DOC concentration reporting. The GE Sievers unit has an internal acidification step; therefore, prior sample manipulation was not required. Changes in DOC concentration over time were used to calculate uptake rates across leachate sources. Major anion concentration samples (collected at the same intervals as DOC concentration) in triplicate were filtered through 0.4 μm 47 mm nucleopore filters and frozen for preservation prior to analysis. Deionized water was used as a filtration blank and samples were analyzed on a Dionex ICS-1100 ion chromatography system. Stoichiometric ratios of DOC and dissolved N and P concentrations were calculated on a molar basis.
Cellular respiration
A Micro-Oxymax closed-circuit respirometer (Columbus Instruments) was used to measure CO2 and oxygen (O2) concentrations. Each leachate extract, standardized for C concentration plus stream water, was run in triplicate, and blank control samples included leachate extracts without stream water. For the purpose of this experiment the production of CO2 was used for subsequent analyses. After the completion of the experiment, blank values for each leachate source were subtracted from the CO2 production values at each time point.
Bacterial cell abundances
Samples for bacterial cell enumeration were preserved with pre-filtered (0.2 μm) formalin to a final concentration of 2% v/v. Bacteria were filtered onto polycarbonate membranes (0.2 μm pore size) and stained with SYBR® Gold (final concentration 25X, Invitrogen). A Nikon E800 epifluorescence microscope was used to count at least 30 randomly-selected fields, with each field containing a minimum of 20 cells per grid (Lisle et al. 2004), resulting in the enumeration of a minimum of 1000 cells. Triplicate filters for each sample were prepared and counted.
Optical spectroscopy
All samples prepared for optical spectroscopy analyses were filtered through 0.2 μm low-carbon filters into clean, combusted amber glass vials. Absorbance spectra were measured with a Genesys 10 s Series Spectrophotometer (ThermoScientific) from 190 to 1100 nm prior to excitation emission matrix (EEMs) analysis. Leachate extracts and experimental samples (day 0–44) were also measured for absorbance values at a wavelength of 254 nm. Values at A = 254 nm were used to calculate the specific UV absorbance (SUVA254), which was normalized to the C concentration of each sample (Weishaar et al. 2003). Samples with A254 > 0.3 absorbance units (a.u.) were diluted with Milli-Q water to prevent inner-filter effects when post-processing the fluorescence spectroscopy data (Miller and McKnight 2010; Miller et al. 2010). Dilution factors generated in this step (e.g., by half, etc.) were then incorporated into the calculations to produce accurate fluorescence data during the EEMs post-processing steps (see below).
A Horiba Jobin–Yvon Fluoromax-4 Spectrofluorometer equipped with a Xe lamp light source and a pathlength of 1 cm at 25 °C was used to generate EEMs. Using a quartz cuvette, samples were placed into the instrument and spectra were obtained with the following specifications: excitation (Ex) wavelengths 240–450 nm scanned over 10 nm intervals, emission (Em) wavelengths 300–560 nm recorded in 2 nm increments, data integration period 0.25 s, 5 nm band pass for both Ex and Em monochromators. All data was generated in signal/reference mode to normalize the Em signal relative to the Ex light intensity. EEMs post-processing was performed in MATLAB to correct for inner-filter effects and Raman scattering. A Milli-Q water blank was run each day and subsequently subtracted during post-processing. EEMs were also collected for the original stream water sample and each leachate extract.
Molecular composition analysis by ESI FT-ICR MS
Samples were prepared for FT-ICR MS with 200 mg Bond Elut PPL Solid Phase Extraction (SPE; Agilent Technologies) cartridges and eluted with HPLC-grade methanol following previously reported procedures (Dittmar et al. 2008) into combusted amber vials, and stored at 4 °C in the dark prior to analyses. Mass spectra were generated by negative ion mode Electrospray Ionization (ESI) coupled to a 9.4 T FT-ICR mass spectrometer at the National High Magnetic Field Laboratory (NHMFL), in Tallahassee, Florida, with the following parameters: flow rate 0.5 μL/min, needle voltage 2.7 kV, tube lens 350 V, heated metal capillary operated at 6 W, Ex range m/z 100–1500 at a frequency sweep rate of 50 Hz/ls, and octopole frequencies were maintained at 2.0 MHz. Multiple (50–200) time domain acquisitions were co-added, Hanning apodized, and zero-filled once before fast Fourier transformation and magnitude calculation (Marshall and Verdun 1990). NHMFL software was used to calibrate the data and generate peak lists for each sample. Molecular formulae were limited to mass spectral peaks calculated to be six times the baseline root mean square noise (6σ), a conservative threshold. Mass spectra were internally calibrated with commonly occurring DOM known methylene (–CH2) homologous series compounds linked across 200–700 Da and the data was sorted for molecular formula assignment by previously established methods (Stenson et al. 2003). Molecular formula composition was assigned manually (detailed descriptions reported in D’Andrilli et al. (2013)) and confirmed with EnviroOrg (NHMFL software by Yuri Corilo © EnviroOrg™, Florida State University: 2016). Elemental composition assignments of DOM included all possible naturally occurring molecular combinations of C, H, N, O, and S, confirmed with individual monoisotopic mass spectral peaks, assignment error < 1 ppm, and homologous series group confirmation (D’Andrilli et al. 2013). Data were post-processed for molecular composition comparative analyses by calculating heteroatom content (percentages of N0–3 and/or S0–1 containing CcHhOo species) and chemical reactivity using the molecular lability boundary (MLB) and lability percentages (MLBL) (D’Andrilli et al. 2015).
DNA extraction, PCR amplification, sequencing, and analysis
Samples for DNA sequencing analysis (stream water and day 2 grass, leaf, and needle incubations) were immediately filtered onto 47 mm Supor®-200, 0.2 μm pore size, sterile membranes under low pressure (< 7 psi). Filters were transferred to cryovials and stored at − 80 °C until further processing. Genomic DNA from biomass collected on filters was extracted using the PowerSoil DNA extraction kit (MoBio, Carlsbad, CA, USA) following manufacturer’s recommendations. Extracted genomic DNA was quantified using the Qubit DNA Assay Kit (Molecular Probes, Eugene, OR, USA). The V3/V4 regions of the 16S rRNA gene were amplified in 50 μL PCR reactions. PCR reactions contained 100–200 pg of extracted genomic DNA, 0.1 μM of each primer (UW Biotechnology Center, Madison, WI, USA), a 1X final concentration of Bulls Eye PREMIUM Taq 2X Mix (Midwest Scientific, St Louis, MO, USA), and nuclease free water. The primer design consisted of the Illumina adaptor sequences followed by either the universal 341F 5′acactctttccctacacgacgctcttccgatctCCTACGGGNGGCWGCAG -3′ or 805R 5′gtgactggagttcagacgtgtgctcttccgatctGACTACHVGGGTATCTAATCC -3′. PCR was performed in an Eppendorf Mastercycler pro S. The amplification protocol consisted of an initial denaturation at 95 °C for 3 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 30 s, and a final extension step at 72 °C for 5 min. A negative PCR control without DNA template added was run in parallel with every PCR sample. The presence of PCR products of the correct size was confirmed by band visualization in a 1.0% agarose Tris–acetate EDTA gel stained with GelRed™.
Paired end, 250 bp sequencing was performed on a Illumina MiSeq Sequencer at the University of Wisconsin-Madison Biotechnology Center, forward and reverse sequences were joined with the Quantitative Insights Into Microbial Ecology (QIIME) toolkit version 1.9.0 (Caporaso et al. 2010). Contigs were subsequently analyzed with the Mothur platform v.1.34.4 (Schloss et al. 2009). For sequence quality refinement, sequences containing ambiguous bases, homopolymers longer than eight bases, or an average quality score below 30 over a 50 bp window, were excluded from further analysis (Schloss et al. 2011). The maximum sequence length was 469 bp, and sequences shorter than 450 bp were removed. Processed sequences were aligned against the SILVA Gold database in Mothur. Chimeric sequences were removed using UCHIME (Edgar et al. 2011) in combination with the SILVA Gold database and a second chimera check using the sequence collection from the present study as a database. Sequences were classified with a Bayesian method (Wang et al. 2007) using the Mothur formatted version of the RDP classifier. Operational taxonomic units (OTUs) were defined at ≥ 97% 16S rRNA sequence identity. Sequences were deposited in the SRA database under the accession number SRP097447. The 200 most abundant microbial OTUs (i.e. those making up ~ 50% of the dissimilarity between samples) responsible for differences in community structure between the original stream water sample and leachate incubation samples at day 2 were analyzed using a similarity percentage (SIMPER) analysis in the statistical package Primer v.6.2 (Clarke and Gorley 2006). Prior to the SIMPER analysis, OTUs were square-root transformed to balance the contributions of rare and common OTUs (Zar 1996).
Statistical analyses
Short- versus long-term processing
Differences in DOM processing through time were assessed using several metrics. DOC concentration decay was assumed to follow a negative exponential decay and therefore a first order rate constant would describe the natural log-transformed change in DOC concentration through time. However, natural log-transformed DOC concentrations showed an apparent non-linear relationship, suggesting the potential for distinct processing dynamics or different DOM “pools” through time. To distinguish these dynamics for each leachate source, we fit models of the natural log-transformed relative DOC concentration (%) over time, with a fixed y-intercept at ln(100%). We also fit a breakpoint regression of a single breakpoint using the ‘segmented’ package in R (Muggeo 2008). We then used AICc model selection to select between the single or two pool model. From this analysis, we used the estimated breakpoint as a temporal boundary for short-term processing. We then compared responses in rates and relative DOC loss and total CO2 accrual among DOM sources at these timescales. Long-term processing was described by the total DOC removed throughout the experiment, a proxy for total lability (sensu Guillemette and del Giorgio 2011), or CO2 accrued.
EEMs multivariate parallel factor (PARAFAC) analysis
Parallel factor (PARAFAC) analysis was used to decompose the complex DOM fluorescence signatures of the EEMs data over all treatments and time points into individual fluorescing regions (Decomposition Routines for Excitation Emission Matrices; drEEM, and the N-way toolbox in MATLAB) (Murphy et al. 2013; Stedmon and Bro 2008). No samples were identified as outliers, a non-negativity constraint was applied to both Ex and Em loadings, and PARAFAC analysis resolved three components (components 1, 2, and 3; C1, C2, and C3) comprising the EEMs data set, explaining 97.5% of the total variance. The PARAFAC model was validated by split-half analysis with all the components of the split model test finding a match with a Tucker correlation coefficient > 0.95 (Murphy et al. 2013). To analyze the contribution of each component compared to individual time points and leachate C sources, percentages of fluorescence maxima [Fmax; Units (R.U.)] for each component within one sample were calculated, e.g., C1 % = [C1 Fmax/(C1 Fmax + C2 Fmax + C3 Fmax)]*100).
DOM processing efficiency
The efficiency of DOM processing through time was calculated by standardizing the DOC uptake rate (U) between sampling time points to mean microbial cell abundances (U ∗ 106 cells−1). The relationship of DOM processing efficiency was then compared to DOM reactive nature, determined from the proportion of EEMs fluorescence at low Ex/Em wavelengths (Cory and Kaplan 2012), and the corresponding Pearson’s correlation coefficient was calculated.
Temporal patterns of DOM composition
To elucidate trends in DOM molecular composition data derived from FT-ICR MS among leachate sources and over time, we used non-metric multidimensional scaling (NMDS), a multivariate statistical technique commonly applied to biological communities. Briefly, NMDS is an ordination technique that constructs a matrix of dissimilarities to graphically represent differences between multiple communities. In this analysis, individual molecular formula (e.g., C18H34N0O8S1) were treated as analogous to biological “species” that were either present or absent within a leachate C source at a given time point. In this manner, FT-ICR MS molecular composition data was prepared for NMDS by creating a unique identification assignment of the individual molecular formulae, for example, a composition assignment of C18H34N0O8S1 is identified by the combination of elements and number of atoms as a text string ID: C18H34N0O8S1. Resemblance matrices using Jaccard similarity index were then used to construct ordinations that allowed for the visualization of similarities and differences among entire “communities” of molecular formulae for leachate C sources over time. Further, commonly derived chemical classes associated with van Krevelen diagrams (e.g., lignin-, tannin-, and protein-like, etc.; see H/C and O/C classification categories in Table S1, as in Smith et al. 2018) as vectors were used to characterize coarse-scale patterns in molecular changes within the leachate C sources throughout the experiment. Vectors representing chemical classes were checked for the normality and “normalized” following protocols outlined by Clarke and Gorley (2006) in Primer 6 (version 6.1.13). Relationships between chemical classes and patterns in molecular formulae over time were examined using Distance based Linear Modeling (DistLM Primer E).
Results
Litter and initial leachate extract characterization
Bulk litter sources differed in quality as determined by elemental content and stoichiometry (Table 1). By these measures, bulk organic matter from grass had the lowest P content and highest C:P and N:P ratios (1398 and 25.6; Table 1), but contained moderate N. Bulk organic matter from leaf was the most replete in P and depleted of N relative to other elements (C:N = 103.2 and N:P = 8.7; Table 1). Needle litter contained the highest N content with a moderate P content.
Leachate extract SUVA254 values ranged from 0.79 to 1.33 L mg−1 m−1, which sit within the spectrum of natural values, but are comparatively low (Weishaar et al. 2003), similar to DOM data collected from other aquatic systems with low vascular plant inputs (Spencer et al. 2008).
EEMs were collected on the original stream water sample, along with the leachate extracts (Fig. S1a–d). Broad fluorescence distributions were only observed for the stream water, corresponding to more complex DOM fluorescence at low and high Ex/Em wavelengths. Stream water EEMs contained maximum fluorescence at Ex: 240–245 nm and Em: 410–440 nm (Fig. S1a), characteristic of fluorophore peak A, which describes more complex chemical species potentially arising from terrestrial plants and/or soils. Grass, leaf, and needle leachate extract EEMs (Fig. S1b–d) contained low Ex/Em wavelength fluorescence only, characteristic of DOM fluorophore peaks B and T, indicative of less complex chemical species. Grass leachate extract contained a maximum fluorescent region at Ex: 260–280 nm and Em: 320–330 nm, characteristic of the fluorophore peak T, and fluorescent maxima for leaf and needle leachate extracts were situated at Ex: 270 nm and Em: 300–310 nm, a region commonly associated with fluorophore peak B. Both fluorophore peak B and T regions describe fluorescent chemical species with low molecular weights and low aromaticity (similar in structure to tyrosine and tryptophan) compared to fluorescence at higher Ex/Em wavelengths. Fluorescence intensities (R.U.) for leachate extracts were greatest for leaf, followed by needle and grass.
The Fluorescence Index (FI) values were variable, spanning 1.33 to 2.04. Leaf and needle leachate extracts were more similar to values reported for microbially derived DOM, whereas the index value for grass was in the range commonly reported for terrestrially DOM (Table 1). The proportion of more-labile leachate extract fluorescence at low Ex/Em wavelengths was greatest for grass, followed by leaf and needle. Leachate extract molecular chemical lability was characterized by the molecular composition percentages of MLBL and was ordered as grass > leaf > needle.
DOC concentration dynamics, cellular respiration, and bacterial cellular abundances
DOC concentrations rapidly decreased within the first few days for all leachate treatments (Fig. 1a, Table S2) and rates of DOC consumption varied among leachate sources in time and in relation to DOC concentration (Fig. 1a inset). Breakpoint analysis of natural log-transformed DOC concentration suggested a breakpoint at ~ 7 days (mean 7.1; range 6.3–7.6 days; Table S3) for all leachate sources. Using this analysis, we classified “short-term” processing to occur between days 0–7, while "long-term" processing was represented by the entirety of the experiment.
Temporal patterns in biological processing of leachate carbon sources (grass, leaf, and needle) throughout 44 days for a natural log-transformed dissolved organic carbon (DOC) concentration (mg/L), b carbon dioxide (CO2) accumulation (µL), and c bacterial cell abundances (cells/mL). The average (dashed line) and range (grey shaded region) of the estimated threshold concluding the short-term processing scale based on breakpoint analysis of DOC concentration data is shown. The relationship between DOC uptake rate (U) and concentration is shown in the inset
Grass DOC concentrations decreased more rapidly than leaf or needle within the first day of the experiment. Notably, the DOC concentration of the leaf amendments showed the slowest rate of DOC consumption within the first day, yet on both short- and long-term scales, maintained the fastest rate of DOC consumption (Fig. 1a). By day 4, the last sampled time point within our short-term processing window, DOC consumed was 32.8%, 49.8%, and 60.8% of original concentrations for needle, grass, and leaf. Within the short-term processing period (0– ~ 7 days), estimated DOC consumption ranged from 57 to 83%, corresponding to k coefficients of − 0.11, − 0.20, and − 0.25 for needle, grass, and leaf.
DOC consumption rates slowed through time to < ~ 1% of their maximum, leading to DOC losses between day 30 and 44 < 3.5% (2.9%, 1.1%, and 3.3% for grass, leaf, and needle leachate treatments). DOC decay rates of the long-term processing period (44 days) were reduced compared to short-term processing, ranging from − 0.005 to − 0.013 across leachate types (mean − 0.01 ± 0.002 1SE; Table S3). After 44 days, the percent of DOC concentration remaining ranged from 11.2% (leaf) to 23.1% (needle) of the original concentration (Fig. 1a; Table S2). Environmental conditions were standardized including initial microbial community structure and biomass, therefore, variation in DOC concentration loss can be directly attributed to chemical differences among DOM sources.
Accumulation of CO2, a direct measure of metabolism linking microbial respiration to leachate source utilization, generally mirrored patterns in DOC consumption through time. Within the short-term processing period, > 60% of total CO2 accumulation had taken place and showed divergent patterns among leachate sources (grass = 385.5 μL, leaf = 521.6 μL, needle = 470 μL). Long-term CO2 patterns differed from short-term patterns and showed an ~ 25% difference in total accumulation among leachate sources (grass = 626.1 μL, needle = 758.5 μL, leaf = 787.4 μL; Fig. 1b). Comparisons of total CO2 accumulation among leachate sources highlighted divergent patterns between DOC decay and CO2 accumulation. In contrast to the patterns of DOC concentration, grass accumulated the least CO2 over the experiment differing from both leaf and needle treatments, which accumulated similar amounts.
Bacterial cell abundances for each leachate treatment at day 0 were within the same order of magnitude (105 cells/mL ± 1.00–3.07 × 104 cells/mL; Fig. 1c, Table S2). Within the short-term scale, bacterial abundances increased 2–3 orders of magnitude in each DOM treatment, with much of this growth occurring by the first day. Over long-term scales, abundances returned to near initial values and remained at these abundances throughout the experiment.
Dissolved nutrient concentrations and stoichiometry showed evidence of fluctuating limitation among leachate sources through time (Table S2, Fig. S2). However, DIN and soluble reactive phosphorous were present at the end of the experiment (with the exception of DIN in the needle treatment) suggesting nutrient limitation does not completely explain the observed dynamics of DOM decomposition.
Bulk DOM fluorescent characterization
Leachate DOM showed a shift in fluorescence signatures from low to high Ex/Em wavelengths over time (Fig. S3). Low Ex/Em wavelength fluorescence, characteristic of amino acid-like fluorophores was present on both short- and long-term processing scales, while higher Ex/Em wavelength fluorescence, characteristic of more degraded/humic-like fluorophores was only present on long-term processing scales. The regions of resolved fluorophores from low to high Ex/Em wavelength fluorescence indicate a gradient of reactive chemical constituents, where Ex: 240–270 nm and Em: 300–350 nm, encompassing fluorophore peaks B and T, correspond to more labile, less complex material when compared with Ex: 240–400 nm and Em: 400–560 nm, characteristic of fluorophore peak regions A, C, and M (Coble et al. 1998, 1990; Cory and Kaplan 2012). Leaf DOM showed the only combination of low and high Ex/Em wavelength fluorescence as separate resolved fluorophore regions within the short-term scale across all leachate C sources. This signature is commonly associated with microbial activity (fluorescence ranging between Ex: 290–325 nm and Em: 370–430 nm) (Fellman et al. 2010 and references within) and may reflect heightened DOM processing for leaf treatment.
PARAFAC analysis identified three fluorescent components (C1, C2, and C3, Fig. 2a). C1, C2, and C3 represented fluorescence typical of “humic-like” terrestrial soil and freshwater processed DOM, “tyrosine-like” freshly produced and degraded DOM, and “tryptophan-like” microbially derived DOM, respectively. PARAFAC component contributions fluctuated with leachate C source and over short- and long-term processing scales (Fig. 2b), with the greatest changes observed from day 0 to 30. Component contribution trends were similar for each C source over time, depicting an increase in C1, decrease in C2, and short-term scale fluctuations followed by a consistent contribution of C3. More complex/degraded fluorescence contributions (C1) increased over time, with grass DOM having the greatest contribution on long-term processing scales. The C2 leaf leachate DOM contribution decreased the fastest on short- and long-term processing scales, capturing the most reactive chemical species over time.
PARAFAC analysis results for dissolved organic matter fluorescence over the 44-day experiment showing a fluorescent components one, two, and three (C1, C2, and C3) of the PARAFAC model including all time points and b the percentage of each component contributing to the overall fluorescence signature per sample over time. Fluorescent data from the PARAFAC model were reported in Raman units. The average (dashed line) and range (grey shaded region) shows the estimated threshold concluding the short-term processing scale based on breakpoint analysis of DOC concentration data. Data for each leachate type is represented with different symbols and colors (b)
DOM processing efficiency
The efficiency by which DOM was processed showed two distinct periods corresponding to the dynamics of microbial communities (Fig. 3). Across all leachate sources, the greatest DOM labile percentages occurred within our short-term window, excluding one measurement, in which bacterial cell abundances and DOC concentrations were highest. The DOM processing efficiency (DOC uptake rate normalized to cell counts) during this time was unrelated to labile DOM percentage. Beyond our estimated short-term processing window, DOM processing efficiency varied ~ tenfold and was highly correlated with fluorescent DOM labile nature percentages (Fig. 3; Pearson’s r = 0.91, p < 0.001).
Relationship between dissolved organic matter (DOM) uptake rate (U) adjusted for cell counts (processing efficiency) and estimated chemical lability among leachate carbon sources across the experiment. DOM lability percentages were calculated by the proportion of fluorescence at low excitation and emission wavelengths from the EEMs data according to Cory and Kaplan (2012). Data point shape corresponds to the carbon leachate source and data point color represents the measured bacterial cellular abundances from high (red) to low (blue). Axis ticks and corresponding numbers along the top axis represent the sampling day of the experiment
DOM FT-ICR MS molecular composition and chemical characterization
Leachate C DOM composition comparisons
The leachate DOM molecular compositions (CcHhNnOoSs) varied in hydrogen saturation, oxygenation, chemical speciation, and reactivity, but showed larger compositional differences through time rather than by C source (Figs. 4, S4). For the FT-ICR MS analysis, only days 0 and 2 fell within our short-term processing scale window, which were selected based on the largest differences in initial and peak biomass growth abundances. Heterogeneity percentages (Table 2) were very similar across leachate C sources at day 0, and grass was the most chemically labile (19.3%), followed by needle (15.2%) and leaf (10.8%) over broad O/C ratios for lipid-, protein-, and amino sugar-/carbohydrate-like chemical categories. Comparatively, leaf DOM contained the most heterogeneous DOM composition at day 0 (highest percentages of CcHhOoN1, CcHhOoN2, CcHhOoN1S1, and CcHhOoN2S1), but at the lowest degree of hydrogen saturation (reflected in the lower chemical lability percentages).
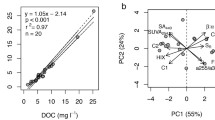
Non-metric multidimensional scaling (NMDS) ordinations of FT-ICR MS molecular composition data among leachate sources for a days 0, 2, and 44, b the short-term processing scale represented by days 0 and 2, and c the long-term processing scale including days 2 and 44. Overlain vectors represent commonly derived chemical classes associated with characteristic H/C and O/C ratios visualized on van Krevelen diagrams (definitions provided in Table S1) and are used for illustrative purposes in (b) and (c). Bolded vectors characterized 78% and 83% of the variation between leachate carbon sources for short- and long-term processing scales, respectively
Heterogeneity decreased by day 2 (concurrent with increasing CcHhOo; Table 2), however, percentages of CcHhOoN2S1 chemical species increased > 3 ×, and chemical lability also increased, consistent with the pattern observed for lability at day 0 among leachate C sources. At day 2, grass DOM was the most heterogeneous, followed by leaf and needle. More similar composition percentages were observed for leaf and needle DOM at day 2.
By day 44, compositional heterogeneity was at its greatest, with the lowest reported percentages for solely CcHhOo containing species. Notably, the greatest contribution of heterogeneous chemical species fluctuated throughout the experiment, from CcHhOoS1 at days 0 and 2, to CcHhOoN1 by day 44. Chemical lability percentages dropped to their lowest values by day 44, describing less hydrogen saturated composition, and followed the same trend observed throughout the DOM experiment (% MLBL grass > needle > leaf). All composition percentages were different at day 44 than at day 0 and the original stream water grab sample (Table 2), reflecting specific changes related to temporal DOM processing. Needle DOM at day 44 also represented the only C source and time point where molecular heterogeneity was also comprised of CcHhOoN3 (5.65%).
Temporal decompositions of the FT-ICR MS DOM composition by NMDS
NMDS ordination revealed distinct shifts in molecular composition through time regardless of leachate C source (Fig. 4a–c). Leachate C sources were separated by a combination of both NMDS axis 1 and 2. Further, visual inspection showed a clear separation between days 0 and 2, and between days 2 and 44 along NMDS axis 2. DOM at day 44 were separated nearly exclusively along NMDS axis 2. More similar molecular compositions among the leachate C sources were observed for day 0 and day 44 along NMDS axis 1, compared to day 2, describing the largest degree of molecular composition differences within our short-term processing scale.
DistLM analysis determined that two DOM chemical classes, lignin-like and unsaturated hydrocarbon-like, explained 78% of the variation between C sources on day 0 and day 2, with both classes more prevalent in day 0 samples (Fig. 4b). Further, three different DOM chemical classes, tannin-, lignin-, and amino sugar-/carbohydrate-like, characterized 83% of the variation between day 2 and day 44, with amino sugar-/carbohydrate-like chemical species being more prevalent at day 2 and tannin- and lignin-like chemical species more prevalent at day 44 (Fig. 4c).
Microbial community assemblages
The raw Illumina MiSeq data set consisted of 229,192 (± 13,119) reads per sample (n = 4; original stream water and day 2 grass, leaf, and needle incubated treatments). Quality refined sequence libraries contained on average 61,769 (± 15,484) reads, which clustered into 1,074 OTUs across all samples. Taxonomic analysis at the class level showed that four heterotrophic lineages dominated across stream water and day 2 leachate C source treatments (Betaproteobacteria, Gammaproteobacteria, Sphingobacteria, and Flavobacteria; Fig. 5a–d top horizontal colored bars) and made up between 92.8 and 99.9% of the microbial population. Comparative panels in Fig. 5a–d show the dominant heterotrophic lineages along with the corresponding DOM molecular composition visualized on van Krevelen diagrams. This approach was used to gain insight between the heterotrophic diversity and microbially produced DOM molecular composition, comparing the starting material (i.e. stream water; Fig. 5a) with the biological and chemical reorganizations after 2 days of incubation for each leachate C source (Fig. 5b–d). The observed number of OTUs showed similar patterns when compared to the predicted alpha diversity indices (Chao and Inv Simpson), indicating that microbial assemblages were sequenced to a sufficient depth (Table S4). The greatest diversity based on Chao richness estimates was found in the original stream water (740.0–745.2; 95% CI). Observed and predicted diversity for each leachate C source decreased at day 2, compared to the stream water. The Chao richness estimates were highest for grass leachate DOM at day 2 (137.0–139.4; 95% CI), followed by needle (109.0–111.8; 95% CI), with leaf supporting the lowest levels of biological diversity (88.1–90.1; 95% CI). This trend was also observed when comparing the heterogeneous nature among leachate C source DOM molecular compositions at day 2, i.e. lower CcHhOo percentages can indicate greater degrees of heterogeneous nature.
Dissolved organic matter (DOM) molecular composition plotted in van Krevelen diagrams and relative abundances of heterotrophic phyla of microbial assemblages for a the collected stream water, and DOM produced by day 2 samples specific to b grass, c leaf, and d needle leachate incubations. The van Krevelen diagrams are plotted as a function of hydrogen to carbon (H/C) and oxygen to carbon (O/C) ratios, calculated from the molecular formulae determined by FT-ICR MS. Gray symbols in the background (b–d) are the molecular formulae common between day 2 C sources and their respective day 0 compositions, whereas the colored symbols in the foreground indicate newly produced material not originally detected at day 0. Relative abundances of microbial assemblages were determined from the MiSeq 16S rRNA gene sequences and percentages are reported at the class taxonomic level
Betaproteobacteria were the dominant lineage in stream water (61%). A decrease in Betaproteobacteria was observed for all leachate C sources after 2 days, with the grass treatment containing much greater abundances (47%) than leaf (17%) and needle treatments (12%). Conversely, Sphingobacteria spp. and Gammaproteobacteria spp. increased for each leachate treatment after 2 days, compared to the stream water assemblages. After 2 days, the leaf and needle incubation microbial populations responded most similarly to one another. Additionally, similar responses of leaf and needle DOM were observed in both newly produced and categorically persistent (i.e. common composition with day 0) molecular composition across H/C and O/C distributions and in heterogeneous nature (Fig. 5b–d). Overall, dissimilarity between microbial OTUs was greatest when comparing grass, leaf, and needle treatments on day 2 to the stream water (Table S5). Grass were > 3 × more dissimilar than needle versus leaf with the greatest differences in Betaproteobacteria, Sphingobacteria, and Flavobacteria abundances.
Discussion
Understanding the temporal dynamics of DOM processing is crucial for discerning the role of C in ecosystem processes (e.g., respiration, flux of CO2 to atmosphere, role in food webs, etc.). While much work has focused on the importance of small and discrete, yet highly biologically reactive pools of DOM, recent studies have begun to highlight the limitations of these viewpoints in relation to the full spectrum of DOM reactivity in freshwater systems (Guillemette and del Giorgio 2011). Our study builds on these ideas by combining a suite of qualitative and quantitative DOM measurements in a controlled experiment to showcase both small and discrete changes of microbial-DOM interactions and their implications for ecosystem processes on a larger scale. Interestingly, the efficiency of processing, bacterial abundances, and chemical composition among leachate C sources all grouped more temporally than among C source material at any given time, highlighting general patterns of DOM consumption and transformation that are independent of source material chemistry. However, differences in the cumulative total and initial rates of CO2 accumulation among leachate C sources may be more dependent on DOM source composition. Therefore, we conclude that composition plays a large role in ecosystem function on short- and long-term scales affecting the mechanisms of processing occurring.
Patterns in DOM processing through time can be highly variable due to shifting intrinsic chemical characteristics and extrinsic biological and environmental variables as DOM is transported and processed within and across ecosystems (Amaral et al. 2016; Smith et al. 2018). Often, the process of DOC decay is modeled assuming first-order kinetics, however, there are a number of theoretical and empirical reasons why DOC loss via decomposition may diverge from an exponential functional form (Adair et al. 2008; Cornwell and Weedon 2014). We observed divergence from an exponential form, potentially arising from shifting biological and/or chemical processes regulating DOM consumption through time. Within the short-term period, we observed an increase in bacterial cell abundances that was similar across all leachate sources. Following these increases, microbial abundances returned to initial levels—three orders of magnitude less. Large swings in microbial biomass might explain, in part, a divergence from first order DOM decay because the abundance and biomass of microbial communities are important components in the rate of DOM uptake and respiration (Wei et al. 2015). This is supported further by the similar bacterial abundance response and similar timing of breakpoints of DOC concentration and respiration shifts in processing among the leachate C sources.
The chemical composition and character of DOM can provide insight into the reactivity of DOM in the environment. Here, the composition of DOM, assessed through both fluorescence and FT-ICR MS, among leachate sources varied both initially and through time. Across leachate sources, the greatest changes in fluorescence contributions corresponded with the greatest changes in DOC concentration. Initial fluorescence was largely composed of tyrosine-like (C2) chemical species, which is often associated with freshly produced and degraded proteinaceous materials (Meyers and Lallier-Verges 1999). Over time, fluorescence from tyrosine-like DOM decreased, which corresponded with a relative increase in more complex/degraded DOM compositions (C1), an expected product of biological processing (Hansen et al. 2016). Notably, the increase in C1 for leaf by day 2 corresponded with the accumulation of DOM having higher Ex/Em wavelengths compared to low Ex/Em wavelength fluorescence signatures observed for days 1–4 (Fig. S3). This signature corresponded to the highest rates of decomposition, likely transitional byproducts of microbial activity, not observed for grass and needle DOM. Importantly, this fluorescence signature, contributing to C1 in the PARAFAC model, describes a different DOM chemistry (Stedmon and Markager 2005; Stedmon et al. 2003), characteristic of degradation (fluorophore peaks A, T, and M contributions) (Fellman et al. 2010) compared to the commonly reported “humic-like” fluorescence of higher plant/soil DOM (Coble 1996). Therefore, this leaf signal (Fig. 3 red triangle below 40% lability), may not be a break from the general trend, but rather represents the caveat within EEMs lability calculations, described by Cory and Kaplan (2012). While bulk level fluorescence data were useful in capturing these general patterns of DOM dynamics they may obscure more refined changes in molecular level identification and chemical characterizations specific to DOM production, transformation, and consumption.
Higher resolution analysis by FT-ICR MS indicated different DOM chemical compositions associated with bacterial production and consumption. Interestingly, molecular composition varied more across time points than among sources, consistent with organic matter biological processing experiments in sediments (Wu et al. 2018). These differences were most pronounced within the DOM molecular composition at day 2, indicating the loss or preferential consumption of DOM constituents within our short-term processing window. At day 2, the decrease in overall heterogeneity coincided with an increase in chemical lability (i.e. hydrogen saturation) over a broad range of oxygenation, which may be associated with N uptake, biomass building, other pathways of microbial metabolism, and potentially non-light induced abiotic chemical reactions (Table 2, Fig. S4).
The dynamic feedback between DOM chemical composition and biological community structure shapes the consumption and transformation of C sources. All biological communities shifted, relative to the original stream water community, towards reduced microbial diversity after 2 days of biological processing. At day 2, microbial diversity was unrelated to DOC consumption. Rather, the leachate source that exhibited the lowest microbial diversity, leaf, corresponded with the largest loss of DOC concentration and the highest accumulation of CO2 at both the time of measurement (day 2) and at the estimated shift in respiration response (day 7). This pattern may reflect two, non-mutually exclusive processes. The local environment chosen to collect the microbial source population is characterized by a riparian community largely composed of aspen, the source for our leaf leachate. Across both terrestrial and aquatic systems, organisms that depend on leaf material as a primary energy source are often adapted to its characteristics. This adaption leads to an apparent ‘home field advantage’ characterized by higher ecological function on local versus transplanted C sources (Ayres et al. 2009; Jackrel and Wootton 2014). Additionally, the DOM chemical composition from leaf leachate (i.e. the specific combination of labile material and the largest percentage of N-containing chemical species at the outset) may allow for greater and more sustained rates of metabolism, intrinsically, or within the nutrient availability of our experiment. Greater percentages of N-containing DOM chemical species have also been linked to greater and more sustained rates of respiration (i.e. largest O2 consumption and CO2 accumulation, comparatively) among microbial and terrestrial end-member C sources for a single microbial isolate (Smith et al. 2018). Therefore, metabolic processes leading to various rates of CO2 accumulation may be dependent on the combination of initial C source composition (including both chemical lability and heterogeneous composition) and the metabolic capacity of biological consortia present.
Predicting the fate of DOM in aquatic ecosystems is an important objective. Initial metrics of DOM quality based on EEMs lability and molecular chemical composition provide insight into its reactive nature but are limited in temporal scale when other biological information is not available. Initial snap shot measurements of lability may only describe patterns of immediate metabolic response and underestimate the complex interactions between DOM heterogeneous composition and microbial community composition over longer-term scales. Within one day, patterns of DOC loss and CO2 accumulation among leachate C sources showed complex relationships with initial quality measures of both particulate and dissolved forms. The grass C leachate was the most labile, as determined by metrics of fluorescence (EEMs lability; Table 1) and FT-ICR MS molecular composition (MLBL; Tables 1 and 2). Grass DOM showed the highest consumption and CO2 production within the first day and was associated with the highest microbial diversity. The higher diversity within the microbial community associated with grass DOM may have initially promoted more rapid microbial metabolism of the more labile grass energy source leading to the unique newly produced molecular composition on short-term scales. However, initial rates of metabolic activity were not sustained. By day 2, the metabolic response associated with leaf DOM had surpassed that of grass DOM and the total long-term function associated with grass leachate by either total DOC loss or CO2 accumulation was 27% and 20% lower than that of leaf DOM, suggesting that varying qualitative aspects of DOM composition may regulate processing over different timescales.
The qualitative characterization metrics of leachate sources through particulate litter C:N:P stoichiometry, and dissolved leachate SUVA254, FI, EEMs lability, and MLBL indicated a qualitative separation of the grass leachate from leaf and needle sources. These source assessments may be related to the similarities observed between leaf and needle leachate molecular composition and microbial community assemblages at day 2. Interestingly, using CO2 accumulation as a metric, both leaf and needle DOM display long-term respiration > 20% of that of grass. However, we observed a divergent pattern in the total DOC lost between leaf and needle leachate that was unexpected based on the similar respiration response (Fig. 1a, b). This discrepancy highlights potentially different conversion processes of DOC to CO2 among leachate C sources and/or microbial communities (Fasching et al. 2014), which also may be related to key chemical differences in the original DOM composition. We did not measure final microbial community composition in our experiment and suggest that future work may benefit from exploring the microbe-C source interactions that regulate the conversion of DOC to CO2.
In the environment, the interwoven nature of physical, chemical, and biological variables confounds our understanding of aquatic DOM processing, ecosystem function, and the DOM or biological signatures useful to predict ecological dynamics. By design, isolated studies simplify away the real complexities of natural systems. Yet, they provide the baseline to probe when, where, and how interactions drive DOM dynamics, i.e. the key factors in the microbial-DOM feedback loop, and raise questions such as: (i) Would a mixed C source approach reflect a simple summation of the processes, degradation products, and average CO2 concentration respired? (ii) How would the respiration patterns change with changing DOM source and microbial consortia along a flow path? Answers to these questions will require a similar experimental design involving both isolated and mixed C source approaches so that linkages among chemical properties, biological communities, and ecosystem function can be directly related to individual processes and microbial-DOM feedback loops in more complex systems.
Conclusions
Our experiment complements recent efforts that are beginning to untangle the importance of the coupled nature of biological processing and chemical energy source for examining the decomposition of DOM at the molecular level in aquatic ecosystems (Fasching et al. 2014; Smith et al. 2018; Wu et al. 2018). The combination of chemical and biological measurements at the peak of biomass growth by day 2 within our short-term period revealed that the patterns of rapid respiration rates and initial CO2 accumulation were a function of microbial community reorganization, rapid DOC concentration loss, and an abundance of labile chemical energy sources. Further, we found changes in DOM composition through time, highlighting byproducts of DOM processing and transformation independent of C source. Patterns within the first few days varied before the conclusion of the short-term timescale but after day 7 were generally consistent with function over longer timescales. Long-term respiration rates were associated with DOM N content and the hydrogen saturated composition of chemical species at the outset; highlighting the dual role of DOM as both an energy and nutrient source. In all, the feedbacks between the chemical character of DOM and biological structure and respiration mediate the production, transformation, and consumption of C in ecosystems. Future work is needed to explore how these feedbacks operate in more complex systems incorporating both isolates and mixtures of diverse C sources and processing time distributions.
References
Adair CE, Parton WJ, Grosso SJD, Silver WL, Harmon ME, Hall SA, Burke IC, Hart SC (2008) Simple three-pool model accurately describes patterns of long-term litter decomposition in diverse climates. Glob Chang Biol 14:2636–2660
Amaral V, Graeber D, Calliari D, Alonso C (2016) Strong linkages between DOM optical properties and main clades of aquatic bacteria. Limnol Oceanogr 61:906–918
Ayres E, Steltzer H, Simmons BL et al (2009) Home-field advantage accelerates leaf litter decomposition in forests. Soil Biol Biochem 41:606–610
Bernhardt ES, Likens GE (2002) Dissolved organic carbon enrichment alters nitrogen dynamics in a forest stream. Ecology 83:1689–1700
Brookshire ENJ, Valett HM, Thomas SA, Webster JR (2005) Coupled cycling of dissolved organic nitrogen and carbon in a forest stream. Ecology 86:2487–2496
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Clarke KR, Gorley RN (2006) PRIMER v6: User Manual/Tutorial. Plymouth Routines in Multivariate Ecological Research, pp. 1–192
Coble PG (1996) Characterization of marine and terrestrial DOM in seawater using excitation emission matrix spectroscopy. Mar Chem 51:325–346
Coble PG, Green SA, Blough NV, Gagosian RB (1990) Characterization of dissolved organic-matter in the Black-Sea by fluorescence spectroscopy. Nature 348:432–435
Coble PG, Del Castillo CE, Avril B (1998) Distribution and optical properties of CDOM in the Arabian Sea during the 1995 Southwest Monsoon. Deep-Sea Res Part II 45:2195–2223
Cole JJ, Prairie YT, Caraco NF et al (2007) Plumbing the global carbon cycle: integrating inland waters into the terrestrial carbon budget. Ecosystems 10:172–185
Cornwell WK, Weedon JT (2014) Decomposition trajectories of diverse litter types: a model selection analysis. Methods Ecol Evol 5:173–182
Cory RM, Kaplan LA (2012) Biological lability of streamwater fluorescent dissolved organic matter. Limnol Oceanogr 57:1347–1360
D’Andrilli J, Foreman CM, Marshall AG, McKnight DM (2013) Characterization of IHSS Pony Lake fulvic acid dissolved organic matter by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry and fluorescence spectroscopy. Org Geochem 65:19–28
D’Andrilli J, Cooper WT, Foreman CM, Marshall AG (2015) An ultrahigh-resolution mass spectrometry index to estimate natural organic matter lability. Rapid Commun Mass Spectrom 29:2385–2401
Dittmar T, Koch B, Hertkorn N, Kattner G (2008) A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater. Limnol Oceanogr-Meth 6:230–235
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Fasching C, Behounek B, Singer GA, Battin TJ (2014) Microbial degradation of terrigenous dissolved organic matter and potential consequences for carbon cycling in brown-water streams. Sci Rep 4:4981
Fellman JB, Hood E, Spencer RGM (2010) Fluorescence spectroscopy opens new windows into dissolved organic matter dynamics in freshwater ecosystems: a review. Limnol Oceanogr 55:2452–2462
Fisher SG, Likens GE (1973) Energy flow in bear brook, new hampshire–integrative approach to stream ecosystem metabolism. Ecol Monogr 43:421–439
Gonsior M, Peake BM, Cooper WT, Podgorski D, D’Andrilli J, Cooper WJ (2009) Photochemically induced changes in dissolved organic matter identified by ultrahigh resolution fourier transform ion cyclotron resonance mass spectrometry. Environ Sci Technol 43:698–703
Guillemette F, del Giorgio PA (2011) Reconstructing the various facets of dissolved organic carbon bioavailability in freshwater ecosystems. Limnol Oceanogr 56:734–748
Hansen AM, Kraus TEC, Pellerin BA, Fleck JA, Downing BD, Bergamaschi BA (2016) Optical properties of dissolved organic matter (DOM): effects of biological and photolytic degradation. Limnol Oceanogr 61:1015–1032
Hotchkiss ER, Hall RO, Sponseller RA, Butman D, Klaminder J, Laudon H, Rosvall M, Karlsson J (2015) Sources of and processes controlling CO2 emissions change with the size of streams and rivers. Nat Geosci 8:696
Jackrel SL, Wootton JT (2014) Local adaptation of stream communities to intraspecific variation in a terrestrial ecosystem subsidy. Ecology 95:37–43
Jaffé R, McKnight D, Maie N, Cory R, McDowell WH, Campbell JL (2008) Spatial and temporal variations in DOM composition in ecosystems: the importance of long-term monitoring of optical properties. J Geophys Res 113:G04032
Lam B, Baer A, Alaee M, Lefebvre B, Moser A, Williams A, Simpson AJ (2007) Major structural components in freshwater dissolved organic matter. Environ Sci Technol 41:8240–8247
Lisle JT, Hamilton MA, Willse AR, McFeters GA (2004) Comparison of fluorescence microscopy and solid-phase cytometry methods for counting bacteria in water. Appl Environ Microbiol 70:5343–5348
Logue JB, Stedmon CA, Kellerman AM, Nielsen NJ, Andersson AF, Laudon H, Lindstrom ES, Kritzberg ES (2016) Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. ISME J 10:533–545
Maie N, Yang C, Miyoshi T, Parish K, Jaffé R (2005) Chemical characteristics of dissolved organic matter in an oligotrophic subtropical wetland/estuarine. Limnol Oceanogr 50:23–35
Marshall AG, Verdun FR (1990) Fourier transforms in NMR, optical, and mass spectrometry: a user’s handbook. Elsevier, Amsterdam
McKnight DM, Boyer EW, Westerhoff PK, Doran PT, Kulbe T, Andersen DT (2001) Spectrofluorometric characterization of dissolved organic matter for indication of precursor organic material and aromaticity. Limnol Oceanogr 46:38–48
Meyers PA, Lallier-Verges E (1999) Lacustrine sedimentary organic matter records of Late Quaternary paleoclimates. J Paleolimnol 21:345–372
Miller MP, McKnight DM (2010) Comparison of seasonal changes in fluorescent dissolved organic matter among aquatic lake and stream sites in the Green Lakes Valley. J Geophys Res 115:G00F12
Miller MP, Simone BE, McKnight DM, Cory RM, Williams MW, Boyer EW (2010) New light on a dark subject: comment. Aquat Sci 72:269–275
Mineau MM, Wollheim WM, Buffam I, Findlay SEG, Hall RO, Hotchkiss ER, Koenig LE, McDowell WH, Parr TB (2016) Dissolved organic carbon uptake in streams: a review and assessment of reach-scale measurements. J Geophys Res 121:2019–2029
Muggeo VMR (2008) segmented: an R package to fit regression models with broken-line relationships. R News 8(1):20–25
Murphy KR, Stedmon CA, Graeber D, Bro R (2013) Fluorescence spectroscopy and multi-way techniques. PARAFAC. Anal Methods 5:6557–6566
Osburn CL, Morris DP, Thorn KA, Moeller RE (2001) Chemical and optical changes in freshwater dissolved organic matter exposed to solar radiation. Biogeochemistry 54:251–278
Pastor A, Catalán N, Nagar N, Light T, Borrego CM, Marcé R (2018) A universal bacterial inoculum for dissolved organic carbon biodegradation experiments in freshwaters. Limnol Oceanogr Methods 16:421–433
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schloss PD, Gevers D, Westcott SL (2011) Reducing the EFfects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6(12):e27310
Seidel M, Dittmar T, Ward ND, Krusche AV, Richey JE, Yager PL, Medeiros PM (2016) Seasonal and spatial variability of dissolved organic matter composition in the lower Amazon River. Biogeochemistry 131:281–302
Smith HJ, Tigges M, D’Andrilli J, Parker A, Bothner B, Foreman CM (2018) Dynamic processing of DOM: insight from exometabolomics, fluorescence spectroscopy, and mass spectrometry. Limnol Oceanogr Lett 3:225–235
Spencer RGM, Aiken GR, Wickland KP, Striegl RG, Hernes PJ (2008) Seasonal and spatial variability in dissolved organic matter quantity and composition from the Yukon River basin, Alaska. Glob Biogeochem Cycles 22:GB002
Stedmon CA, Bro R (2008) Characterizing dissolved organic matter fluorescence with parallel factor analysis: a tutorial. Limnol Oceanogr 6:572–579
Stedmon CA, Markager S (2005) Resolving the variability in dissolved organic matter fluorescence in a temperate estuary and its catchment using PARAFAC analysis. Limnol Oceanogr 50:686–697
Stedmon CA, Markager S, Bro R (2003) Tracing dissolved organic matter in aquatic environments using a new approach to fluorescence spectroscopy. Mar Chem 82:239–254
Stenson AC, Marshall AG, Cooper WT (2003) Exact masses and chemical formulas of individual Suwannee River fulvic acids from ultrahigh resolution electrospray ionization Fourier transform ion cyclotron resonance mass spectra. Anal Chem 75:1275–1284
Vannote RL, Minshall GW, Cummins KW, Sedell JR, Cushing CE (1980) River continuum concept. Can J Fish Aquat Sci 37:130–137
Vonk VE, Tank SE, Mann P, Spencer JR, Treat CC, Striegl RG, Abbott BW, Wickland KP (2015) Biodegradability of dissolved organic carbon in permafrost soils and aquatic systems: a meta-analysis. Biogeosciences 12:6915–6930
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wei H, Chen X, Xiao G, Guenet B, Vicca S, Shen W (2015) Are variations in heterotrophic soil respiration related to changes in substrate availability and microbial biomass carbon in the subtropical forests? Sci Rep 5:18370
Weishaar JL, Aiken GR, Bergamaschi BA, Fram MS, Fujii R, Mopper K (2003) Evaluation of specific ultraviolet absorbance as an indicator of the chemical composition and reactivity of dissolved organic carbon. Environ Sci Technol 37:4702–4708
Wetzel RG (1995) Death, detritus, and energy flow in aquatic ecosystems. Freshw Biol 33:83–89
Wu X, Wu L, Liu Y et al (2018) Microbial interactions with dissolved organic matter drive carbon dynamics and community succession. Front Microbiol 9:1234
Zar JH (1996) Biostatistical analysis, 3rd edn. Prentice-Hall, London
Acknowledgements
This work was supported by the National Science Foundation Division of Materials Research through DMR-1157490 for FT-ICR MS analyses at the National High Magnetic Field Laboratory (NHMFL) in Tallahassee, Florida, USA, as well as the Montana Academy of Sciences Student Research Grant Program awarded to H.J.S., and the Center for Biofilm Engineering at Montana State University through C.M.F. We thank Dr. David C. Podgorski and the Ion Cyclotron Resonance Facility Staff members at the NHMFL for FT-ICR MS support, Melissa Basham for her encouragement and support of the experiment, and Dr. Robert A. Payn for his intellectual creativity and suggestions to improve this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Breck Bowden.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
D’Andrilli, J., Junker, J.R., Smith, H.J. et al. DOM composition alters ecosystem function during microbial processing of isolated sources. Biogeochemistry 142, 281–298 (2019). https://doi.org/10.1007/s10533-018-00534-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10533-018-00534-5