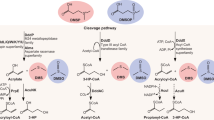
Abstract
Bacterial species associated with the dimethylsulfoniopropionate (DMSP)-producing phytoplankton Scrippsiella trochoidea were cultured and identified, with the aim of establishing their ability to metabolise DMSP, dimethylsulfide (DMS) and dimethylsulfoxide (DMSO). Results demonstrate that of the cultivable bacteria only α-Proteobacteria were capable of producing DMS from DMSP. The concentration of DMSP was shown to affect the amount of DMS produced. Lower DMSP concentrations (1.5 μmol dm−3) were completely assimilated, whereas higher concentrations (10 μmol dm−3) resulted in increasing amounts of DMS being produced. By contrast to the restricted set of bacteria that metabolised DMSP, ~ 70% of the bacterial isolates were able to ‘consume’ DMS. However, 98-100% of the DMS removed was accounted for as DMSO. Notably, a number of these bacteria would only oxidise DMS in the presence of glucose, including members of the γ-Proteobacteria and Bacteroidetes. The observations from this study, coupled with published field data, identify DMS oxidation to DMSO as a major transformation pathway for DMS, and we speculate that the fate of DMS and DMSP in the field are tightly coupled to the available carbon produced by phytoplankton.




Similar content being viewed by others
References
Archer SD, Gilbert FJ, Nightingale PD, Zubkov MV, Taylor AH, Smith GC, Burkill PH (2002) Transformation of dimethylsulphoniopropionate to dimethyl sulphide during summer in the North Sea with an examination of key processes via a modelling approach. Deep-Sea Res II 49(15):3067–3101
Azam F (1998) Microbial control of oceanic carbon flux: the plot thickens. Science 280(5364):694–696
Bell WH, Lang JM, Mitchell R (1974) Selective stimulation of marine bacteria by algal extracellular products. Limnol Oceanogr 19(5):833–839
Biebl H, Pukall R, Lunsdorf H, Schulz S, Allgaier M, Tindall BJ, Wagner-Dobler I (2007) Description of Labrenzia alexandrii gen. nov., sp. nov., a novel alphaproteobacterium containing bacteriochlorophyll a, and a proposal for reclassification of Stappia aggregata as Labrenzia aggregata comb. nov., of Stappia marina as Labrenzia marina comb. nov. and of Stappia alba as Labrenzia alba comb. nov., and emended descriptions of the genera Pannonibacter, Stappia and Roseibium, and of the species Roseibium denhamense and Roseibium hamelinense. Int J Syst Evol Microbiol 57(5):1095–1107. doi:10.1099/ijs.0.64821-0
Charlson RJ, Lovelock JE, Andreae MO, Warren SG (1987) Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 326:655–661
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucl Acids Res 37(Suppl 1):D141–D145. doi:10.1093/nar/gkn879
Curson AR, Rogers R, Todd JD, Brearley CA, Johnston AW (2008) Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate-changing gas dimethylsulfide in several marine a-proteobacteria and Rhodobacter sphaeroides. Environ Microbiol 10(3):757–767
Dalton H, Stirling DI, Quayle JR (1982) Co-metabolism [and discussion]. Phil Trans R Soc Lond B 297(1088):481–496. doi:10.1098/rstb.1982.0056
de Souza MP, Yoch DC (1995) Comparative physiology of dimethyl sulfide production by dimethylsulfoniopropionate lyase in Pseudomonas doudoroffii and Alcaligenes sp. strain M3A. Appl Environ Microbiol 61(11):3986–3991
del Valle DA, Kieber DJ, Kiene RP (2007) Depth-dependent fate of biologically-consumed dimethylsulfide in the Sargasso Sea. Mar Chem 103(1–2):197–208
del Valle DA, Kieber DJ, Toole DA, Brinkley J, Kiene RP (2009) Biological consumption of dimethylsulfide (DMS) and its importance in DMS dynamics in the Ross Sea. Antarctica. Limnol Oceanogr 54(3):785–798
DeLong EF, Franks DG, Alldredge AL (1993) Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr 38(5):924–934
deZwart JMM, Nelisse PN, Kuenen JG (1996) Isolation and characterization of Methylophaga sulfidovorans sp nov: An obligately methylotrophic, aerobic, dimethylsulfide oxidizing bacterium from a microbial mat. FEMS Microbiol Ecol 20(4):261–270
Fandino LB, Riemann L, Steward GF, Long RA, Azam F (2001) Variations in bacterial community structure during a dinoflagellate bloom analyzed by DGGE and 16S rDNA sequencing. Aquat Microb Ecol 23(2):119–130
Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S (2006) Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA 103(35):13104–13109
Fuse H, Ohta M, Takimura O, Murakami K, Inoue H, Yamaoka Y, Oclarit JM, Omori T (1998) Oxidation of trichloroethylene and dimethyl sulfide by a marine Methylomicrobium strain containing soluble methane monooxygenase. Biosci Biotechnol Biochem 62(10):1925–1931
González JM, Kiene RP, Moran MA (1999) Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl Environ Microbiol 65(9):3810–3819
Green DH, Llewellyn LE, Negri AP, Blackburn SI, Bolch CJS (2004) Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol Ecol 47(3):345–357
Green DH, Hart MC, Blackburn SI, Bolch CJS (2010) Bacterial diversity of Gymnodinium catenatum and its relationship to dinoflagellate toxicity. Aquat Microb Ecol 61(1):73–87. doi:10.3354/ame01437
Green DH, Shenoy D, Hart MC, Hatton AD (2011) DMS oxidation coupled to biomass production by a marine Flavobacterium. Appl Environ Microbiol. doi:10.1128/AEM.02675-10
Hatton A, Wilson S (2007) Particulate dimethylsulphoxide and dimethylsulphoniopropionate in phytoplankton cultures and Scottish coastal waters. Aquatic Sci: Res Across Boundar 69(3):330–340
Hatton AD, Malin G, McEwan AG, Liss PS (1994) Determination of dimethyl sulfoxide in aqueous solution by an enzyme-linked method. Anal Chem 66(22):4093–4096. doi:10.1021/ac00094a036
Hatton AD, Turner SM, Malin G, Liss P (1998) Dimethylsulphoxide and other biogenic sulphur compounds in the Galapagos plume. Deep Sea Res II 45(6):1043–1053
Hatton AD, Darroch L, Malin G (2004) The role of dimethylsulphoxide in the marine biogeochemical cycle of dimethylsulphide. Oceanogr Mar Biol Annu Rev 42:29–56
Horinouchi M, Kasuga K, Nojiri H, Yamane H, Omori T (1997) Cloning and characterization of genes encoding an enzyme which oxidizes dimethyl sulfide in Acinetobacter sp strain 20B. FEMS Microbiol Lett 155(1):99–105. doi:10.1111/j.1574-6968.1997.tb12692.x
Howard EC, Henriksen JR, Buchan A, Reisch CR, Burgmann H, Welsh R, Ye W, Gonzalez JM, Mace K, Joye SB, Kiene RP, Whitman WB, Moran MA (2006) Bacterial taxa that limit sulfur flux from the ocean. Science 314(5799):649–652
Howard EC, Sun S, Biers EJ, Moran MA (2008) Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ Microbiol 10(9):2397–2410
Keller MD, Bellows WK, Guillard RRL (1989) Dimethylsulfide production in marine phytoplankton. In: Saltzman ES, Cooper WJ (eds) Biogenic sulfur in the environment. American Chemical Society, Washington, pp 167–181
Kiene RP, Linn LJ (2000a) Distribution and turnover of dissolved DMSP and its relationship with bacterial production and dimethylsulfide in the Gulf of Mexico. Limnol Oceanogr 45(4):849–861
Kiene RP, Linn LJ (2000b) The fate of dissolved dimethylsulfoniopropionate (DMSP) in seawater: tracer studies using 35S-DMSP. Geochim Cosmochim Acta 64(16):2797–2810
Kiene RP, Linn LJ, Bruton JA (2000) New and important roles for DMSP in marine microbial communities. J Sea Res 43(3–4):209–224
Ledyard KM, Dacey JWH (1996) Microbial cycling of DMSP and DMS in coastal and oligotrophic seawater. Limnol Oceanogr 41(1):33–40
Liss PS, Hatton AD, Malin G, Nightingale PD, Turner SM (1997) Marine sulphur emissions. Philosophical Transactions- Royal Society of London Series B Biological Sciences 352:159–167
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar BuchnerA, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H (2004) ARB: a software environment for sequence data. Nucl Acids Res 32(4):1363–1371
Malmstrom RR, Kiene RP, Kirchman DL (2004) Identification and enumeration of bacteria assimilating dimethylsulfoniopropionate (DMSP) In the North Atlantic and Gulf of Mexico. Limnol Oceanogr 49(2):597–606
Miller TR, Hnilicka K, Dziedzic A, Desplats P, Belas R (2004) Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl Environ Microbiol 70(8):4692–4701
Mitchell JG, Okubo A, Fuhrman JA (1985) Microzones surrounding phytoplankton form the basis for a stratified marine microbial ecosystem. Nature 316:58–59
Rinta-Kanto JM, Bürgmann H, Gifford SM, Sun S, Sharma S, del Valle DA, Kiene RP, Moran MA (2011) Analysis of sulfur-related transcription by Roseobacter communities using a taxon-specific functional gene microarray. Environ Microbiol 13(2):453–467. doi:10.1111/j.1462-2920.2010.02350.x
Scarratt M, Cantin G, Levasseur M, Michaud S (2000) Particle size-fractionated kinetics of DMS production: where does DMSP cleavage occur at the microscale? J Sea Res 43(3–4):245–252
Schäfer H (2007) Isolation of Methylophaga spp. from marine dimethylsulfide-degrading enrichment cultures and identification of polypeptides induced during growth on dimethylsulfide. Appl Environ Microbiol 73(8):2580–2591
Simó R (2001) Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol Evol 16(6):287–294
Simó R, Hatton AD, Malin G, Liss PS (1998) Particulate dimethyl sulphoxide in seawater: production by microplankton. Mar Ecol Prog Ser 167:291–296
Simó R, Archer SD, Pedrós-Alió C, Gilpin L, Stelfox-Widdicombe CE (2002) Coupled dynamics of dimethylsulfoniopropionate and dimethylsulfide cycling and the microbial food web in surface waters of the North Atlantic. Limnol Oceanogr 47(1):53–61
Stefels J (2000) Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J Sea Res 43(3–4):183–197
Sunda W, Kieber DJ, Kiene RP, Huntsman S (2002) An antioxidant function for DMSP and DMS in marine algae. Nature 418:317–320
Todd JD, Rogers R, Li YG, Wexler M, Bond PL, Sun L, Curson ARJ, Malin G, Steinke M, Johnston AWB (2007) Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315(5812):666–669. doi:10.1126/science.1135370
Todd JD, Curson ARJ, Dupont CL, Nicholson P, Johnston AWB (2009) The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environ Microbiol 11(6):1376–1385
Todd JD, Curson AR, Kirkwood M, Sullivan MJ, Green RT, Johnston AW (2011) DddQ, a novel, cupin-containing, dimethylsulfoniopropionate lyase in marine roseobacters and in uncultured marine bacteria. Environ Microbiol 13(2):427–438. doi:10.1111/j.1462-2920.2010.02348.x
Tripp HJ, Kitner JB, Schwalbach MS, Dacey JWH, Wilhelm LJ, Giovannoni SJ (2008) SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452(7188):741–744
Turner SM, Malin G, Liss PS, Harbour DS, Holligan PM (1988) The seasonal variation of dimethyl sulfide and dimethylsulfonioproprionate concentrations in nearshore waters. Limnol Oceanogr 33(3):364–375
Turner SM, Malin G, BÂgander LE, Leck C (1990) Interlaboratory calibration and sample analysis of dimethyl sulphide in water. Mar Chem 29:47–62
van Duyl FC, Gieskes WWC, Kop AJ, Lewis WE (1998) Biological control of short-term variations in the concentration of DMSP and DMS during a Phaeocystis spring bloom. J Sea Res 40(3–4):221–231
Vila-Costa M, del Valle DA, Gonzalez JM, Slezak D, Kiene RP, Sanchez O, Simo R (2006) Phylogenetic identification and metabolism of marine dimethylsulfide-consuming bacteria. Environ Microbiol 8(12):2189–2200
Vila-Costa M, Rinta-Kanto JM, Sun S, Sharma S, Poretsky R, Moran MA (2010) Transcriptomic analysis of a marine bacterial community enriched with dimethylsulfoniopropionate. Isme J 4(11):1410–1420. doi:ismej201062
Weber CF, King GM (2007) Physiological, ecological, and phylogenetic characterization of Stappia, a marine CO-oxidizing bacterial genus. Appl Environ Microbiol 73(4):1266–1276
Zeyer J, Eicher P, Wakeham SG, Schwarzenbach RP (1987) Oxidation of dimethylsulfide to dimethylsulfoxide by phototrophic purple bacteria. Appl Environ Microbiol 53(9):2026–2032
Zimmer-Faust RK, Souza MPd, Yoch DC (1996) Bacterial chemotaxis and its potential role in marine dimethylsulfide production and biogeochemical sulfur cycling. Limnol Oceanogr 41(6):1330–1334
Zubkov MV, Fuchs BM, Archer SD, Kiene RP, Amann R, Burkill PH (2001a) Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ Microbiol 3(5):304–311
Zubkov MV, Fuchs BM, Burkill PH, Amann R (2001b) Comparison of cellular and biomass specific activities of dominant bacterioplankton groups in stratified waters of the Celtic Sea. Appl Environ Microbiol 67(11):5210–5218
Zubkov MV, Fuchs BM, Archer SD, Kiene RP, Amann R, Burkill PH (2002) Rapid turnover of dissolved DMS and DMSP by defined bacterioplankton communities in the stratified euphotic zone of the North Sea. Deep-Sea Res II 49(15):3017–3038
Zubkov MV, Linn LJ, Amann R, Kiene RP (2004) Temporal patterns of biological dimethylsulfide (DMS) consumption during laboratory-induced phytoplankton bloom cycles. Mar Ecol Prog Ser 271:77–86
Acknowledgments
We are grateful for support from Debra Brennan for 16S rRNA clone analysis, Elaine Mitchell for flow cytometric analysis and Christine Campbell for supply of S. trochoidea CCAP 1134/1. This research was supported by the Natural Environment Research Council UK SOLAS Programme (NE/C51725X/1) and a Natural Environment Research Council-CASE studentship (NE/F006748/1).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Hatton, A.D., Shenoy, D.M., Hart, M.C. et al. Metabolism of DMSP, DMS and DMSO by the cultivable bacterial community associated with the DMSP-producing dinoflagellate Scrippsiella trochoidea . Biogeochemistry 110, 131–146 (2012). https://doi.org/10.1007/s10533-012-9702-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10533-012-9702-7