Abstract
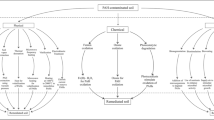
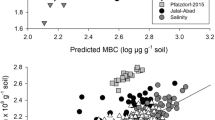
A previous bioremediation survey on a creosote-contaminated soil showed that aeration and optimal humidity promoted depletion of three-ringed polycyclic aromatic hydrocarbons (PAHs), but residual concentrations of four-ringed benzo(a)anthracene (B(a)A) and chrysene (Chry) remained. In order to explain the lack of further degradation of heavier PAHs such as four-ringed PAHs and to analyze the microbial population responsible for PAH biodegradation, a chemical and microbial molecular approach was used. Using a slurry incubation strategy, soil in liquid mineral medium with and without additional B(a)A and Chry was found to contain a powerful PAH-degrading microbial community that eliminated 89% and 53% of the added B(a)A and Chry, respectively. It is hypothesized that the lack of PAH bioavailability hampered their further biodegradation in the unspiked soil. According to the results of the culture-dependent and independent techniques Mycobacterium parmense, Pseudomonas mexicana, and Sphingobacterials group could control B(a)A and Chry degradation in combination with several microorganisms with secondary metabolic activity.


Similar content being viewed by others
References
Abalos A, Viñas M, Sabaté J, Manresa MA, Solanas AM (2004) Enhanced biodegradation of casablanca crude oil by a microbial consortium in presence of a rhamnolipid produced by Pseudomonas aeruginosa AT10. Biodegradation 15:249–260. doi:10.1023/B:BIOD.0000042915.28757.fb
Alexander M (1995) How toxic are toxic chemicals in soil? Environ Sci Technol 29:2713–2717. doi:10.1021/es00011a003
Alexander M (1999) Biodegradation and bioremediation. Academic Press, San Diego
Fanti F, Tortoli E, Hall L, Roberts GD, Kroppenstedt RM, Dodi I, Conti S, Polonelli L, Chezzi C (2004) Mycobacterium parmense sp. nov. Int J Syst Evol Microbiol 54:1123–1127. doi:10.1099/ijs.0.02760-0
Farmer PB, Singh R, Kaur B, Sram RJ, Binkova B, Kalina I, Popov TA, Garte S, Taioli E, Gabelova A, Cebulska-Wasilewska A (2003) Molecular epidemiology studies of carcinogenic environmental pollutants: effects of polycyclic aromatic hydrocarbons (PAHs) in environmental pollution on exogenous and oxidative DNA damage. Mutat Res 544:397–402. doi:10.1016/j.mrrev.2003.09.002
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hareland W, Crawford RL, Chapman PJ, Dagley S (1975) Metabolic function and propierties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J Bacteriol 121:272–285
Maidak BL, Cole JR, Lilburn TG, Parker CT Jr, Saxman PR, Stredwick JM, Garrity GM, Li B, Olsen GJ, Pramanik S, Schmidt TM, Tiedje JM (2000) The RDP (Ribosomal Database Project) continues. Nucleic Acids Res 28:173–174. doi:10.1093/nar/28.1.173
Miller CD, Child R, Hughes JE, Benscai M, Der JP, Sims RC, Anderson AJ (2007) Diversity of soil mycobacterium isolates from three sites that degrade polycyclic aromatic hydrocarbobns. J Appl Microbiol 102:1612–1624. doi:10.1111/j.1365-2672.2006.03202.x
Sabaté J, Viñas M, Solanas AM (2006) Bioavailability assessment and environmental fate of PAHs in biostimulated creosote-contaminated soil. Chemosphere 63:1648–1659. doi:10.1016/j.chemosphere.2005.10.020
Thierry S, Macarie H, Iizuka T, Geissdoerfer W, Assih EA, Spanevello M, Verhe F, Thomas P, Fudou R, Monroy O, Labat M, Ouattara AS (2004) Pseudoxanthomonas mexicana sp. nov. and Pseudoxanthomonas japonensis sp. nov., isolated from diverse environments, and emended descriptions of the genus Pseudoxanthomonas Finkmann et al. 2000 and of its type species. Int J Syst Evol Microbiol 54:2245–2255. doi:10.1099/ijs.0.02810-0
Turenne CY, Tschetter L, Wolfe J, Kabani A (2001) Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J Clin Microbiol 39:3638–3648. doi:10.1128/JCM.39.10.3638-3648.2001
Viñas M, Sabaté J, Solanas AM (2005a) Culture-dependent and -independent approaches establish the complexity of a PAH-degrading microbial consortium. Can J Microbiol 51:897–909. doi:10.1139/w05-090
Viñas M, Sabaté J, Espuny MJ, Solanas AM (2005b) Bacterial community dynamics and PAHs degradation during bioremediation of heavily creosote-contaminated soil. Appl Environ Microbiol 71:7008–7018. doi:10.1128/AEM.71.11.7008-7018.2005
Walter U, Beyer M, Klein J, Rehm HJ (1991) Degradation of pyrene by Rhodococcus sp. UW1. Appl Microbiol Biotechnol 34:671–676. doi:10.1007/BF00167921
Weon HY, Kim BY, Hong SB, Joa HJ, Nam SS, Lee KH, Kwon SW (2007) Skermanella aerolata p. nov., isolated from air, emended description of the genus Skermanella. Int J Syst Evol Microbiol 57:1539–1542. doi:10.1099/ijs.0.64676-0
Wilson SC, Jones KC (1993) Bioremediation of PAHs contaminated soils. Environ Pollut 81:229–249. doi:10.1016/0269-7491(93)90206-4 (Review)
Wrenn BA, Venosa AD (1996) Selective enumeration of aromatic and aliphatic hydrocarbon-degrading bacteria by a most-probable-number procedure. Can J Microbiol 42:252–258
Xie CH, Yokota A (2005) Reclassification of Alcaligenes latus strains IAM 12599T and IAM 12664 and Pseudomonas saccharophila as Azohydromonas lata gen. nov., comb. Nov., Azohydromonas australica sp. nov. and Pelomonas saccharopila gen. nov., comb. Nov., respectively. Int J Syst Evol Microbiol 55:2419–2425. doi:10.1099/ijs.0.63733-0
Yu Z, Morrison M (2004) Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol 70:4800–4806. doi:10.1128/AEM.70.8.4800-4806.2004
Acknowledgments
This study was financially supported by the Spanish Ministry of Science and Technology (CTM2007-61097/TECNO) and by the Reference Network in Biotechnology (XERBA) of the Autonomous Government of Catalonia. The authors declare that the experiments discussed in this paper were performed in compliance with current Spanish and European legislation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lladó, S., Jiménez, N., Viñas, M. et al. Microbial populations related to PAH biodegradation in an aged biostimulated creosote-contaminated soil. Biodegradation 20, 593–601 (2009). https://doi.org/10.1007/s10532-009-9247-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-009-9247-1