Abstract
Genetic tools can have a key role in informing conservation management of declining populations. Genetic diversity is an important determinant of population fitness and resilience, and can require careful management to ensure sufficient variation is present. In addition, population genetics data reveal patterns of connectivity and gene flow between locations, enabling mangers to predict recovery and resilience, identify areas of local adaptation, and generate restoration plans. Here, we demonstrate a conservation genetics approach to inform restoration and management of the loggerhead sponge (Spheciospongia vesparium) in the Florida Keys, USA. This species is a dominant, habitat-forming component of marine ecosystems in the Caribbean region, but in Florida has suffered numerous mass mortality events. We developed microsatellite markers and used them to genotype sponges from 14 locations in Florida and a site each in The Bahamas, Belize and Barbuda. We found that genetic diversity levels were similar across all sites, but inbreeding and bottleneck signatures were present in Florida. Populations are highly structured at the regional scale, whilst within Florida connectivity is present in a weak isolation by distance pattern, coupled with chaotic genetic patchiness. Evidence of a weak barrier to gene flow was found in Florida among sites situated on opposite sides of the islands in the Middle Keys. Loggerhead sponge populations in Florida are vulnerable in the face of mass mortalities due to low connectivity with other areas in the region, as well as distance-limited and unpredictable local connectivity patterns. However, our discovery of Florida’s high genetic diversity increases hope for resilience to future perturbations. These results provide valuable insight for sponge restoration practice in Florida.
Similar content being viewed by others
Introduction
Population declines in keystone species have a number of negative impacts on associated communities, ecosystem functioning, and the provision of ecosystem services (Sweeney et al. 2004; Hicke et al. 2012; Thomson et al. 2015; Sorte et al. 2017). Genetic factors are a significant determinant of population health and fitness, and can influence both the longevity of populations and the success of conservation strategies (Frankham 2005). However, genetic information is unavailable for the vast majority of species, and thus the application of conservation genetics theory to practice has been limited (Shafer et al. 2015; Taylor et al. 2017).
Declining populations are vulnerable to low genetic diversity due to the effects of genetic drift, in which rare alleles have a higher probability of being lost due to random chance in smaller populations. These effects are amplified considerably in populations that experience a rapid decline, or bottleneck, through which substantial genetic variation is randomly eliminated in a short space of time (Sbordoni et al. 1986; Bellinger et al. 2003; Bristol et al. 2013). This threatens population survival, as genetic diversity is an important determinant of long-term population persistence (Frankham 2005). Indeed, high genetic diversity bolsters the resilience of populations, because they harbour a higher adaptive capacity with which to respond to perturbations such as disease, environmental change, or declining environmental conditions (Hughes and Stachowicz 2004; Ehlers et al. 2008; Evans et al. 2017). Low genetic diversity is also related to inbreeding depression, where recessive deleterious alleles are more likely to combine within individuals and reduce fitness (Whitlock 2000; Reed and Frankham 2003; Charlesworth et al. 2009), further compromising the long-term prospects for survival of the population.
Connectivity—the movement of individuals or propagules among populations—is an important counterforce against declining population size, low genetic diversity and local extinction. A well-connected population receives a regular supply of immigrants, thus boosting population size. Crucially, if these migrants successfully reproduce, they can help replenish the gene pool with new alleles, thus countering the effects of genetic drift through gene flow (Garant et al. 2007; Saenz-Agudelo et al. 2011; Frankham 2015). Conversely, isolated populations with little connectivity are more vulnerable to extinction due to limited immigration and gene pool restriction (van der Meer et al. 2013). Assessing levels of genetic connectivity among geographical sites is therefore another key step in managing vulnerable populations.
An interesting case study to apply such genetic information to conservation practice exists among sponge populations in Florida Bay and the Florida Keys (USA). In nearshore hard-bottom habitats in this area, sponges form a dominant component of benthic communities (Chiappone and Sullivan 1994; Tellier and Bertelsen 2008), and perform a number of vital functional roles and ecosystem services. Given their high relative biomass, they provide the majority of architectural complexity and habitat structure in the area (Herrnkind et al. 1997). This is especially important given that Florida Bay is a nursery area for a number of economically important fish and invertebrate species, including snapper (Lutjanus spp.), stone crabs (Menippe mercenaria), and Caribbean spiny lobsters (Panulirus argus). Several species of sponge are themselves the target of commercial fisheries in the region (Butler et al. 2017). Moreover, sponge endosymbionts are important in creating soundscapes that form an acoustic cue for larval settlement in a variety of taxa (Butler et al. 2016). As filter feeders, sponges drive nutrient cycling dynamics in the area (Fiore et al. 2017; Hoer et al. 2018; Valentine and Butler 2019), and contribute to the maintenance of water quality (Peterson et al. 2006; Butler et al. 2018).
Sponge communities in the Florida Keys have suffered a number of mass mortality events (Butler et al. 1995; Stevely et al. 2010; Wall et al. 2012) associated with recurring blooms of the cyanobacteria Synechococcus spp. (Fourqurean and Robblee 1999; Berry et al. 2015), as well as stochastic cold weather events (Colella et al. 2012) and storm damage (Stevely et al. 2010). These mass mortalities have had dramatic consequences for the ecosystem, including declines in local juvenile lobster populations (Butler et al. 1995; Herrnkind et al. 1997), increased susceptibility to further cyanobacterial blooms (Peterson et al. 2006; Wall et al. 2012) and diminished underwater soundscapes predicted to impact larval recruitment from a variety of taxa (Butler et al. 2016). Furthermore, sponge population recovery is potentially forestalled by limited dispersal, as adults are sessile, and sponge larvae are generally short-lived, with larval durations of a few hours to a few days before settlement (Maldonado 2006; Maldonado and Riesgo 2008).
Due to their keystone role in the ecosystem and the impacts of their decline, sponge restoration work has been undertaken in the area for a number of years, where healthy sponges have been fragmented and translocated to areas that have suffered mortalities (Butler et al. 2016; Valentine and Butler 2019). However, cyanobacterial blooms and sponge mass mortalities continue to recur across different areas of the Florida Keys and Florida Bay. Coupled with work to identify the proximal causes of sponge mortality and the implementation of habitat improvement measures, understanding the genetic status of the populations is imperative for future restoration and management planning. In addition, investigating connectivity patterns will aid understanding of source-sink interactions across the Bay, and identify priority areas for restoration.
In this study, we investigated these topics in the loggerhead sponge, Spheciospongia vesparium (Lamarck, 1815). Spheciospongia vesparium is common throughout Florida Bay and has the largest biomass of all sponge species in the Bay (Tellier and Bertelsen 2008). It is also found on reefs and in lagoons throughout the Greater Caribbean region. Reproduction and larval biology have not yet been studied in S. vesparium, therefore limiting our ability to predict dispersal and population genetic pattners. However, studies of other Clionaidae species suggest that varied reproductive characteristics exist within the family: sexual and asexual reproduction have both been observed (Rosell and Uriz 2002; Schönberg 2002; Maldonado and Riesgo 2008), and similarly, gonochorism and hermaphroditism are also both found within the family (Piscitelli et al. 2011; González-Rivero et al. 2013). The Clionaidae are oviparous (i.e., broadcast spawning of both the sperm and eggs) (Ereskovsky 2018), and fertilization and larval development are mainly external, although in Cliona vermifera eggs are fertilized internally and the zygote released (Bautista-Guerrero et al. 2014). Larvae are lecithotrophic (i.e., do not feed), and larval duration is short—in Cliona viridis, it was estimated at < 10 days (Mariani et al. 2000). Clionaidae larvae have so far been observed to show weak swimming ability, with crawling behaviour common (Mariani et al. 2000, 2001).
Here, we aimed to describe patterns of genetic diversity and genetic connectivity in S. vesparium at hard bottom sites across the Florida Keys. In addition, we sampled three other locations in the Greater Caribbean to act as comparative populations, and to observe drivers of population structure at the regional scale.
Methods
Sample collection and preservation
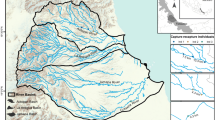
We collected S. vesparium samples from shallow water sites (< 2 m depth) in four main localities: the Florida Keys/ Florida Bay (USA), Abaco Island (The Bahamas), Barbuda, and Caye Caulker (Belize) (Fig. 1; Table 1). We sampled a number of sites across the Upper, Middle and Lower Florida Keys: 12 sites on the Florida Bay side of the Keys and 2 collection sites on the Atlantic side (Table 1; Fig. 1). Our sites in Florida included both those that have previously been affected by cyanobacterial blooms and mortalities, and those that have not. At each site in Florida we sampled between 10 and 32 individuals (average of 18.6 ± 1.2 SEM), and in Abaco, Barbuda, and Caye Caulker we sampled 12, 20, and 10 individuals, respectively (Table 1). We avoided sites where restoration work had taken place in order to observe the natural patterns of population structure and genetic diversity as far as possible. We collected small tissue fragments (~ 2 cm3) and immediately transferred the samples into 95% ethanol, which was renewed after 24 h.
Spheciospongia vesparium sampling sites. Inset map shows sampling sites in Greater Caribbean (BZ: Caye Caulker, Belize; BH: Abaco, The Bahamas; BAR: Codrington Lagoon, Barbuda). Main map shows Florida Keys sampling sites (PK: Pigeon Key; SCB: Snake Creek Basin; SB: Steamboat Channel; CKA: Craig Key (Atlantic); FK: Fiesta Key; LKB: Long Key (Bay-side); LKA: Long Key (Atlantic); GKB: Grassy Key Bank; BK: Bamboo Key; KC: Kemp Channel; LC: Little Crane Key; WK: Waltz Key; BC: Boca Chica Channel; LP: Lakes Passage). Lower Keys = dark purple; Middle Keys = medium pink; Upper Keys = light pink. Basemaps: Natural Earth, ESRI
Microsatellite development
For this study, we characterised twelve new tri- and tetra-nucleotide microsatellite loci (see Supplementary Material for full details of the methods). In brief, DNA from a single S. vesparium sample collected from Long Key (Bay-side) was sequenced using Illumina MiSeq 2 × 250 base pair technology. We then processed the sequence reads using the Palfinder Galaxy bioinformatics pipeline (Griffiths et al. 2016) to quality filter the data, screen for microsatellites and design primers. We tested 36 loci, of which 12 could be successfully amplified and scored, and were subsequently used in this study.
DNA extraction and genotyping
We checked sponge tissue samples under a dissecting microscope to remove any visible endosymbiotic invertebrates, and then extracted total DNA using the DNeasy® Blood and Tissue Kit (Qiagen). We combined 10 of the 12 microsatellite primer pairs in two multiplex (5-plex) PCRs using the fluorophores 6-FAM and HEX (Table S1), and ran two primer pairs, Vesp36 and Vesp9 in singleplex PCRs due to problems encountered in multiplexing these loci. We utilized a three-primer universal tail approach for fluorescent labelling PCR products, as described in Blacket et al. (2012) and Culley et al. (2013). We carried out PCRs using the Type-it® Microsatellite PCR Kit (Qiagen) with the following cycling conditions: 95 °C for 5 min, 28 × (95 °C for 30 s, 60 °C for 90 s, 72 °C for 30 s), 60 °C for 30 min. For any amplification failures, PCRs were repeated in singleplex reactions with lowered (50–59 °C) annealing temperatures.
We sized PCR products by capillary electrophoresis using a 3730 DNA Analyzer (Thermo Fisher Scientific) with GeneScan™ 500, 600 or 1200 LIZ® size standard (Thermo Fisher Scientific), or a homemade ROX-based size standard. On all plates, we included both positive and negative controls. We scored alleles using Genemapper® v3.7 software (Thermo Fisher Scientific), and corrected allele sizes according to the positive controls to account for differences in allele length based on the machine or size standard used. We then binned alleles using the R package ‘MsatAllele’ v1.02 (Alberto 2009) in RStudio v3.0.3 (R Core Team 2014).
Quality control and summary statistics
We calculated the probability of linkage disequilibrium between pairs of loci using Genepop on the Web v4.2 (Raymond and Rousset 1995; Rousset 2008), with p values corrected for multiple tests using the false discovery rate procedure of Benjamini and Yekutieli (2001), as calculated using the R function p.adjust. We estimated null allele frequency at each locus using the EM algorithm (Dempster et al. 1977) in FreeNA (Chapuis and Estoup 2007). As null alleles can cause overestimation of FST values and levels of population differentiation (Chapuis and Estoup 2007), we conducted a post hoc test to test the extent of any bias, as follows: We calculated average null allele frequencies for all loci, and calculated global FST with and without ENA correction for null alleles (as implemented in FreeNA). We then removed the locus with the highest null allele frequency and recalculated the uncorrected and corrected global FST values. We repeated this systematically until just one locus remained. This allowed us to observe the cumulative effects of each locus and their null allele loads on FST by comparing the corrected and uncorrected values.
Genetic diversity, inbreeding and bottlenecks
We used Genodive v2.032b (Meirmans and Van Tiendener 2004) to calculate observed heterozygosity (HO) and gene diversity/ expected heterozygosity (HS; Nei 1987). We also tested for probability of departure from Hardy–Weinberg Equilibrium (HWE) in Genodive using the AMOVA (least squares) method and 50,000 permutations (p values corrected for multiple tests using the Benjamini and Yekutieli method, calculated as previously). We calculated average allelic richness and private allele richness rarefied to the lowest sample size (maximum g = 10) in ADZE v1.0 (Szpiech et al. 2008). We repeated these analyses with all the Florida sites grouped as one population and each separately.
We estimated inbreeding coefficients (Avg Fi) in INEst v2.1 (Chybicki and Burczyk 2009), correcting for the presence of null alleles. The program includes three possible parameters that can affect inbreeding coefficient estimation: null alleles (‘n’), inbreeding (‘f’) and genotyping failure (‘b’). We ran the individual inbreeding model (IMM) for all combinations of these parameters and calculated the Deviance Information Criteria (DIC) for each run to determine the best model fit for the data. We ran the model using 500,000 Monte Carlo Markov Chain (MCMC) cycles with 50,000 burnin cycles.
We used INEst to find evidence of genetic signatures of recent population bottleneck events. The program implements two tests; the first identifies heterozygosity excesses in respect to allelic richness (Cornuet and Luikart 1996), and the second identifies M-ratio (mean ratio of allelic richness to allelic size range) deficiencies (Garza and Williamson 2001). Both phenomena have been observed when populations experience rapid reductions in size. We used the two-phase mutation model, and tested significance using a Wilcoxon signed-rank test with 1000 permutations.
Genetic connectivity patterns
We estimated genetic differentiation among sites by calculating pairwise FST (Wright 1943, 1949) and D (Jost 2008) in Genodive v2.032b. For FST values, we tested their significance in Genodive using 50,000 permutations, and corrected significance for multiple tests using Benjamini-Yekutieli correction as described above.
We used two different approaches to infer the number of population clusters (‘K’) in the data. Firstly, we used the Bayesian individual-based assignment model implemented in the ‘Geneland’ package v.4.0.8 (Guillot et al. 2005; Guillot et al. 2008) in RStudio, which uses spatial and genetic data to infer K and calculate the probability of individual assignment. Due to the assumptions of this model, we used only the seven loci that did not deviate from HWE in the majority of the sites, and deleted samples in which missing data was present in the majority of the HWE loci (n = 285). We first ran the no-admixture model to obtain estimates of cluster membership and allele frequencies. We used the uncorrelated allele frequencies, spatial and null allele models, and ran the program with 1,000,000 MCMC iterations, 100 thinning and 1000 burnin, and uncertainty on coordinates set to 0.0005. We set the maximum number of nuclei to 855, and the maximum rate of the Poisson process to 285. We repeated this with K ranging from 1 to 17, with 10 independent runs for each value of K. We then ran the admixture model using the estimates obtained from the no-admixture run with the highest average posterior probability. For the admixture model, we used 1,000,000 MCMC iterations, a thinning of 100, and a burnin of 1000. We extracted the q-matrix of estimated individual membership proportions to each of the detected clusters, and used Distruct v1.1 (Rosenberg 2004) to graphically display the results.
We used Flock v3.1 (Duchesne and Turgeon 2012) as an alternative method to infer membership to population clusters. This method estimates K and partitions samples into K clusters based on iterated reallocation, uses no a priori information on sampling location, and does not assume populations are in HWE. We tested K from 1 to 17 in 50 independent runs per value of K, and ran each model with 20 iterations (i.e. 20 rounds of reallocation). We used plateau analysis based on log likelihood difference (LLOD) scores, as described by Duchesne and Turgeon (2012), to infer the most likely value of K. We carried out hierarchical clustering approaches for both the Geneland and Flock analyses by first running the models using all sites, and then repeating the process on any multi-site clusters identified.
We used Discriminant Analysis of Principle Components (DAPC) (Jombart et al. 2010) as implemented in the package ‘adegenet’ v.2.1.1 (Jombart 2008) in RStudio to examine genetic variation among the sites based on allele frequencies. We used the function optim.a.score to calculate the optimum number of principle components (PCs) to retain in the analysis to prevent over-fitting of the model, whilst preserving the maximum discriminability. We included all sites in the first instance, and then conducted a further analysis on the Florida sites alone to examine the presence of fine-scale structure.
We carried out a Principle Coordinates Analysis (PCoA) in GenAlEx v6.502 (Peakall and Smouse 2012) using null allele corrected pairwise FST values as calculated previously. We carried out the analysis first on all sites, and then on only the Florida sites. We repeated the analysis using Jost’s D to confirm robustness of the results.
We used an analysis of molecular variance (AMOVA) to examine the partitioning of genetic variation within and among individuals and sites. We included a grouping factor for the Florida sites in order to estimate variation among sites within Florida, and among Florida, The Bahamas, Barbuda and Belize within the analysis. We carried out the AMOVA in Genodive v3.0.0 using the infinite allele model.
We looked for evidence of barriers to gene flow among the Florida and The Bahamas sites using the software Barrier v2.2 (Manni et al. 2004). We excluded the Barbuda and Belize sites from this analysis because of the large geographic distances separating them from the other sites, as this does not offer an appropriate theoretical framework to search for oceanographic barriers. Barrier uses the spatial coordinates of the sampling sites and Delauney triangulation to partition geographic space into polygons, creating a Voronoï tessellation map with each site contained within a single polygon whose edges border neighbouring adjacent sites. Monmonier's (1973) maximum difference algorithm then uses this tessellation map along with a genetic distance matrix (Jost’s D) to detect genetic discontinuities among neighbouring sites. We assessed the robustness of the computed barriers by repeating analysis on 100 resampled bootstrap D matrices. We created the resampled bootstrap matrices in the R package ‘diveRsity’ v1.9.90 (Keenan et al. 2013). We computed increasing numbers of barriers until bootstrap support fell below 50%, reaching a maximum of three barriers. Following computation of barriers, we used AMOVAs to examine the partitioning of genetic variation across barriers.
To test the presence of genetic isolation by distance (IBD) within Florida, we performed a Mantel test to detect association between matrices of linearised pairwise genetic distances (FST/[1 − FST]) and the logarithm of geographic distances. We calculated least-cost oceanographic distances between sites (i.e., the shortest distance possible, excluding landmasses) using ‘marmap’ v0.9.5 (Pante and Simon-Bouhet 2013) in RStudio, and carried out the Mantel tests in ‘ade4’ v1.7-10 (Dray et al. 2007) in RStudio, with 9999 permutations to calculate significance.
We used Geneclass2 v2 (Piry et al. 2004) to detect first generation migrants among the sampling locations, and their putative population origins. We used the Bayesian criteria of Rannala and Mountain (1997) for likelihood estimation, and the Monte Carlo method of Paetkau et al. (2004) for probability computation, with the Lhome criterion, as source populations for all individuals were unlikely to have been sampled. We used a significance threshold of p < 0.01 and carried out simulations with 10,000 individuals.
Results
Quality control and summary statistics
In total, we collected samples from 326 individuals across 17 sites (Table 1). Twenty-two samples were removed from the final dataset due to amplification failure in over 50% of the loci, leaving 304 individuals. Two individuals from the Lakes Passage had identical genotypes, one of which was removed from the dataset for analysis, yielding 303 individuals. Following correction for multiple tests, no significant linkage disequilibrium was found between pairs of loci. Null allele frequency was high in some markers (Tables S1, S2); however, post hoc analysis showed that the null allele-corrected global FST value was only marginally higher (+ 0.002) than the uncorrected value when all loci were included in the analysis (Table S2). Furthermore, the difference between the uncorrected and corrected FST did not increase as more loci were added (r2 = − 0.03608, p = 0.4504), and therefore all loci were retained for the population genetics analyses. The number of alleles per locus over all sites ranged from 4 (Vesp23) to 27 (Vesp30) (Table S1).
Genetic diversity, inbreeding and bottlenecks
Genetic diversity (allelic richness and gene diversity) was slightly lower at the Florida and Barbuda sites compared to The Bahamas and Belize. However, overlapping error bars among many of the sites indicate that this is only significant for a few Florida sites (Table 2; Fig. 2; Table S3). Broadly, genetic diversity can therefore be considered to be the same across Florida and non-Florida sites. Average rarefied allelic richness ranged from 3.408 (Pigeon Key) to 4.399 (Long Key—Bay-side) and gene diversity (HS) ranged from 0.569 (in Pigeon Key) to 0.735 (in Belize) (Fig. 2; Table 2; Table S3).
Observed heterozygosity over all loci varied from 0.251 (Craig Key—Atlantic) to 0.504 (The Bahamas) (Table 2; Table S3). All sites had lower than expected levels of heterozygosity (Table 2; Table S3), and significant departures from HWE were found in a number of loci and populations (Table S4). The DIC analysis in INEst determined either the ‘nfb’ (null allele, inbreeding and genotyping failure) or ‘nb’ (null allele and genotyping failure) models to be the best fit for the sites in this study (Table 2). This indicates that null alleles and genotyping failure would affect inbreeding coefficient estimations in all the sites, but in ten of the sites, inbreeding was also an influential component of the model. The null allele-corrected inbreeding coefficients were positive in all locations, ranging from 0.036 (The Bahamas) to 0.343 (Craig Key—Atlantic). However, the posterior 95% probability intervals included zeros at all sites, and therefore FIS cannot be considered to be significantly above zero. When the Florida sites were grouped together as a single population, however, the posterior 95% probability interval was above zero, which may indicate significant inbreeding across the area.
We found deficiencies in M-ratios at four sites, indicating the presence of recent bottleneck events (Boca Chica Channel, p = 0.0385; Little Crane Key, p < 0.001; Craig Key (Atlantic, p < 0.001; Long Key (Atlantic), p < 0.001). However, none of the sites showed significant heterozygote excess in comparison to allelic richness.
Genetic connectivity patterns
Pairwise FST ranged from -0.019 (no differentiation) between Craig Key (Atlantic) and Long Key (Atlantic), to 0.273 (great differentiation) between Pigeon Key and Barbuda (Table 3). Among the four regional locations (Florida, The Bahamas, Barbuda, Belize), FST values were large and significant, showing strong differentiation. Among sites within Florida, FST values were lower (≤ 0.116), but significant differentiation was present between many pairs of sites. In general, higher differentiation could be observed among Upper and Lower Keys sites than comparisons involving the Middle Keys sites, but patchiness can be observed throughout. Patterns of D were similar, and ranged from -0.035 (between the Craig Key and Long Key Atlantic sites, as previously) to 0.668 (between The Bahamas and Waltz Key) (Table 3). Private alleles were present at many sites (Table 2; Table S3), and average private allelic richness was higher among the non-Florida sites (Table 2; Fig. 2).
Using Geneland, K = 4 was found for each independent run, with each regional location forming a separate population cluster (Fig. 3). In contrast, Flock showed strong evidence for K = 2. Samples were broadly partitioned into a Florida cluster and a cluster comprising individuals from Belize, Barbuda and The Bahamas. Two individuals from Florida (Craig Key Atlantic and Lakes Passage) fell into the Belize, Barbuda and The Bahamas cluster; otherwise, clustering was concurrent with sampling locations. When a second Flock analysis was carried out on the Barbuda, Belize and The Bahamas cluster, the samples were partitioned into K = 3 concurrent with sampling locations. When repeating the models for only the Florida samples, the Geneland model was unable to converge, indicating that K = 1 or the presence of strong isolation by distance in the data. Similarly, no plateau was obtained in Flock, indicating K = 1.
Membership coefficients per individual at K = 4 clusters inferred from Geneland (admixture model) for sponges collected from 14 locations in Florida and a single location each in Barbuda, The Bahamas, and Belize. Individual sponges are each represented by a single bar; colours indicate cluster identity, and height of the bar shows estimated proportion of membership to cluster
The DAPC showed clear separation of the Barbuda and The Bahamas sites from all other sites (Fig. 4a). All the Florida sites clustered together, with inertia ellipses showing substantial overlap among sites. The Belize site clustered closely to the Florida sites, with some Belize samples showing overlap with the Florida point cloud. In the Florida-only DAPC analysis, no clustering patterns were present, but points from sites more closely situated geographically tended to be closer together in the DAPC plot (Fig. 4b).
Discriminant analysis of principle components (DAPC) for S. vesparium sampling sites over a all sites, b Florida sites. Points represent individual sponges, sampling sites are coded by colour, and inertia ellipses summarise the point cloud for each site. Insets are scree plots showing the proportion of principle components retained in the analysis and the proportion of variance they represent (shaded portion)
In the PCoA carried out on all sites (Fig. 5a), the first axis separated Florida from The Bahamas, Belize and Barbuda, and the second separated the Upper Keys from the Lower Keys and Atlantic side of the Middle Keys; the Bay side Middle Keys were distributed among both. In the Florida-only PCoA (Fig. 5b), points were distributed in a loose isolation by distance fashion, but notably the sites on the Atlantic side of the Middle Keys (Long Key and Craig Key) were clustered with Waltz Key, and separated from the sites on the Bay side of the Middle Keys. When the analysis was replicated with Jost’s D instead of FST, the patterns observed were very similar (data not shown).
Principle coordinates analysis (PCoA) using null allele corrected pairwise FST values a among all sampling sites, and b among Florida sites (Upper Keys = light pink, Middle Keys = medium pink, Lower Keys = dark purple). BZ: Caye Caulker, Belize; BH: Abaco, The Bahamas; BAR: Codrington Lagoon, Barbuda; PK: Pigeon Key; SCB: Snake Creek Basin; SB: Steamboat Channel; CKA: Craig Key (Atlantic); FK: Fiesta Key; LKB: Long Key (Bay-side); LKA: Long Key (Atlantic); GKB: Grassy Key Bank; BK: Bamboo Key; KC: Kemp Channel; LC: Little Crane Key; WK: Waltz Key; BC: Boca Chica Channel; LP: Lakes Passage
The AMOVA showed that 18.2% of the total variation was found among the four main locations, while 2.4% was found among the sites within Florida. 30.9% of variation was found among individuals within sites, while 48.5% was within individuals (Table 4).
Barrier software suggested the presence of two barriers with high bootstrap support: the first was a barrier between Florida and The Bahamas, with a bootstrap score of 100%. A second barrier separated the Atlantic sites from their adjacent Bay-side sites in the Middle Keys in Florida (Fig. 6). As barriers are computed based on the tessellation map, this barrier comprised a number of polygon edges, which showed bootstrap support ranging 39–99% (Fig. 6). A further barrier was estimated to separate the Middle and Lower Keys sites, however, this had low bootstrap support (10–54%) (Fig. 6). AMOVA analysis of sites separated by barriers showed that a large proportion of genetic variation was present between Florida and The Bahamas (20.1% of genetic variation, FST = 0.201), while only a small amount of genetic variation was found between the Atlantic and adjacent Bay-side sites in the Middle Keys (3.3% of genetic variation, FST = 0.033) (Table 4). In both cases, more variation was found across the barrier than among sites on the same side of the barrier (Table 4).
a Inferred barriers to gene flow among Spheciospongia vesparium sites in the Florida Keys, using Monmonier’s (1973) algorithm as implemented in Barrier v2.1 software. Green circles represent spatial projection of sites, blue lines show Voronoï polygons, and red lines show inferred barriers. Grey numbers show bootstrap score (out of 100). PK: Pigeon Key; SCB: Snake Creek Basin; SB: Steamboat Channel; CKA: Craig Key (Atlantic); FK: Fiesta Key; LKB: Long Key (Bay-side); LKA: Long Key (Atlantic); GKB: Grassy Key Bank; BK: Bamboo Key; KC: Kemp Channel; LC: Little Crane Key; WK: Waltz Key; BC: Boca Chica Channel; LP: Lakes Passage. b Satellite map of sites at Long Key (Bay-side) (LKB), Long Key (Atlantic) (LKA), Fiesta Key (FK) and Craig Key (Atlantic) (CKA), with red line to show separation of sites by inferred barrier. Basemap: Bing
Isolation by distance within Florida was significant, but the effect size was relatively small (r = 0.229, p = 0.031) (Fig. 7). We obtained comparable results when repeating the analysis with Jost’s D (r = 0.225, p = 0.033).
Three putative first generation migrants were detected. All potential migrants were found within Florida sites, and all originated from other Florida sites. Two migrants were found at Long Key (Bay), with origins from Waltz Key (p = 0.0007; distance 89 km) and Fiesta Key (p = 0.0019, distance 5 km), and the third migrant was found at Little Crane Key with inferred origins of Kemp Channel (p < 0.0001, distance 12 km).
Discussion
Genetic diversity and bottlenecks
Genetic diversity was similar throughout all of the sites sampled in Florida and the Caribbean, and was comparable to levels observed in other demosponge species (Chaves-Fonnegra et al. 2015; Riesgo et al. 2019). This implies that recurring mass mortality events have not significantly reduced genetic diversity in Florida, however, pre-mortality data is not available to confirm this hypothesis. Nevertheless, this study does provide a baseline with which future assessments of genetic diversity can be compared. We did not find signatures of genetic bottlenecks in sites that have been affected by cyanobacterial blooms. However, bottleneck signatures were present in four Florida sites that have not been affected by cyanobacterial blooms. These sites may have suffered unrecorded mortality events due to a different cause, such as disease, climatic variation, or hurricane disturbance. As sponges rapidly disappear once dead, and leave no visible skeleton, mass mortalities in sponges can be overlooked unless specific, regular monitoring is undertaken (Wulff 2006).
Levels of genetic diversity (allelic richness, gene diversity) in other sponge populations that have experienced mass mortalities vary by species. Spongia officinalis has high genetic diversity with no bottleneck signatures (Dailianis et al. 2011), whilst the opposite was found for a congener, S. lamella (Pérez-Portela et al. 2015). In Ircinia fasciculata, evidence of bottlenecks have been found at many (but not all) sites known to have suffered mortalities (Riesgo et al. 2016). In other species within the Florida reef tract, such as the coral Acropora cervicornis and sea urchin Diadema antillarum, genetic diversity was similar to other Caribbean sites tested (Chandler et al. 2017; Drury et al. 2017), despite mass mortality events. High genetic diversity, despite recent mass mortalities, may be due to high levels of connectivity with other sites. This would provide a pathway for re-colonisation and would increase the effective population size (Ne), protecting the population against the effects of genetic drift (Dailianis et al. 2011; Riesgo et al. 2016). However, high variance in reproductive success can occur in broadcast spawning marine invertebrates, reducing Ne, and thus increasing vulnerability to bottlenecks (Hedgecock 1994).
Similarity in genetic diversity across all sampling sites implies that S. vesparium in Florida may still have sufficient genetic variation for resilience against some future stressors. In the Florida Keys, those include anthropogenic effects on water quality and global climate change (Wall et al. 2012; Kearney et al. 2015; Butler and Dolan 2017), as well as further cyanobacteria blooms. However, bottleneck signatures in some sites suggest that genetic diversity may have been previously lost due to unknown causes, and therefore caution should be exercised in management to prevent possible reductions in genetic variation.
Inbreeding and null alleles
Observed heterozygosity was lower in Florida and Barbuda than Belize and The Bahamas. However, all sites showed excesses in homozygosity, and departures from Hardy Weinberg equilibrium were present across loci and sites. This phenomenon can be caused by inbreeding, but it can also be attributed to the presence of null alleles, which were found in a number of loci and across all sampling sites in our study. Null alleles are caused by mutations in primer binding regions that prevent primers from binding, and subsequently cause amplification failure in PCR, either in both alleles (resulting in missing data), or for only one allele (resulting in false homozygotes). High null allele frequencies are commonly found in sponge microsatellite studies (Dailianis et al. 2011; Guardiola et al. 2012, 2016; Chaves-Fonnegra et al. 2015; Pérez-Portela et al. 2015; Richards et al. 2016), suggesting that the problem may be common in the phylum, and is a known issue in other taxa (e.g., molluscs and insects; Chapuis and Estoup 2007). To reduce the impact of null allele bias on our estimates of inbreeding, we corrected FIS values for null alleles.
We found positive FIS values in all populations when corrected for null alleles, but this was not statistically significant in any of the sites when tested individually, potentially due to small sample sizes. Our genetic clustering analyses concluded that Florida was a single population, which enabled us to group the Florida sites for more statistical power, resulting in a significant positive mean FIS value. This suggests the presence of inbreeding in Florida S. vesparium populations, although the large 95% posterior probability intervals at the individual site level preclude a more fine-scale spatial assessment. Inbreeding has negative implications for fitness, thus our results highlight a potential concern for the health, reproductive success and longevity of S. vesparium in Florida.
Inbreeding is often characteristic of populations that have experienced declines. Hence, the positive FIS values we observed for S. vesparium in Florida may be due to mass mortality events, coupled with limited regional-scale connectivity to replenish the gene pool. However, high inbreeding coefficients are widespread in the Porifera (Guardiola et al. 2012; Bell et al. 2014; Chaves-Fonnegra et al. 2015; Pérez-Portela et al. 2015; Giles et al. 2015; Padua et al. 2017). This suggests that the positive FIS values for Porifera may be general characteristics of the phylum, perhaps associated with high philopatry due to limited larval dispersal. Additionally, sponges in the Clionaidae family can be simultaneously hermaphroditic (contain both eggs and sperm at the same time) (Piscitelli et al. 2011), so self-fertilisation, or selfing, could theoretically be possible; however there is currently no recorded evidence of selfing in the Porifera phylum.
Positive FIS values can also be caused by excess homozygosity driven by Wahlund effects. These effects can occur when there is population structure within a site or group, and can be caused by reproductive asynchronicity or recruitment of different genetic cohort (Duran et al. 2004; Chaves-Fonnegra et al. 2015; Riesgo et al. 2016). With this in mind, it is difficult to fully gauge the implications of positive FIS for population health.
Genetic connectivity patterns
Spheciospongia vesparium exhibited strong population structure at the regional (Caribbean) spatial scale, indicating that connectivity among sponge populations in the four countries we sampled is low. These results are congruent with those of other sponge species, which exhibit high differentiation at large spatial scales in the Caribbean (López-Legentil and Pawlik 2009; Chaves-Fonnegra et al. 2015; de Bakker et al. 2016; Richards et al. 2016; DeBiasse et al. 2016), but also in other regions (Duran et al. 2004; Xavier et al. 2010; Pérez-Portela et al. 2015; Riesgo et al. 2016, 2019; Brown et al. 2017; Padua et al. 2017; Taboada et al. 2018). Dispersal in marine species is affected by a number of factors and the complex interactions between them (Cowen et al. 2006; Cowen and Sponaugle 2009), including ocean current patterns and life history characteristics such as pelagic larval duration, larval behaviour, and reproductive strategies (Butler et al. 2011; Selkoe and Toonen 2011; Kough and Paris 2015; Coelho and Lasker 2016). Although reproductive and larval traits for S. vesparium are not known, sponge larvae generally have short pelagic larval durations, limiting their dispersal capacity. This includes previously studied members of the Clionaidae family (Warburton 1966; Mariani et al. 2000, 2001), to which S. vesparium belongs. Furthermore, Clionaidae larvae have been found to exhibit weak swimming ability, and commonly crawl (Mariani et al. 2000, 2001), further minimizing dispersal capacity.
Our results on the population structure of S. vesparium are consistent with expectations of connectivity as determined by regional ocean current patterns. Our analyses indicated the presence of a barrier to gene flow between the Florida sites and Abaco in The Bahamas, which concurs with patterns found in genetic studies of other sponges (López-Legentil and Pawlik 2009; Richards et al. 2016; DeBiasse et al. 2016) and corals (Brazeau et al. 2005; Baums et al. 2010), and biophysical modelling predictions of fish and lobster larvae (Cowen et al. 2006; Truelove et al. 2017). This break is likely due to the strong Florida Current, which runs between The Bahamas and Florida, and can act as a strong barrier to dispersal.
Genetic differentiation was much larger between Florida and Abaco than between Florida and Belize, despite the geographic distance being much larger for the latter pair, as shown by genetic distance calculations and the DAPC analysis. Connectivity between Florida and Belize could be aided by the Caribbean Current and Loop Current, which can support larval transport from Belize towards Florida in as little as 7 to 10 days (Muhling et al. 2013). This is likely to be higher than the larval duration of most sponge species, but locations ‘upstream’ from Florida, such as the Yucatán Peninsula, could act as intermediate ‘stepping stones’ to aid gene flow between these areas, as appears the case for some marine diseases (Kough et al. 2015), thus reducing genetic differentiation.
Connectivity across the Florida Keys
Florida formed a single genetic cluster in our analyses (Geneland, Flock, DAPC), and the AMOVA showed that only 2.4% of the total genetic variation in the dataset was among sites in Florida. These results indicate that some level of connectivity is present across the Keys. We also found evidence of recent migration between Florida sites in our first generation migration analysis. According to genetic distance and PCoA analyses, sites such as Long Key (Bay-side) in the Middle Keys and Boca Chica Channel in the Lower Keys appeared well-connected to sites throughout the Florida Keys range. The complex currents found across the Florida Keys are likely to aid in connectivity among disparate sites. Although the main current dominating the area is the north-easterly running Florida Current, there are many local oceanographic processes that can affect larval dispersal patterns. Westerly running counter currents arise as a result of downwelling winds and offshore eddies and gyres (Lee and Williams 1999; Yeung et al. 2001; Kourafalou and Kang 2012), and eddies themselves also drive connectivity in the area (Sponaugle et al. 2005). Connectivity is also influenced by a species’ life history. Reproduction of S. vesparium has not been described, however, oviparity occurs in some members of the Clionaidae family (Maldonado and Riesgo 2008; González-Rivero et al. 2013); if S. vesparium is also oviparous, additional dispersal of the gametes before fertilization may increase connectivity over longer distances compared to viviparous sponges. However, in other oviparous sponges, egg masses have been observed to stick to the substrate close to the mother sponge due to their envelopment in an adhesive material (Mariani et al. 2001). Furthermore, fertilization rates generally decrease over increasing gametic dispersal distances in broadcast spawners (Levitan 1991; Lauzon-Guay and Scheibling 2007). However, even a relatively small proportion of far-dispersing eggs that get successfully fertilized could increase the genetic connectivity between populations (Trakhtenbrotl et al. 2005).
Despite evidence of connectivity, there was still population structure among the Florida sites, demonstrating that the area does not form a completely panmictic population. Isolation by distance accounted for some of the structure across sites: genetic similarity decreases with geographic distance. This suggests that distance-limited dispersal influences population structure on smaller (< 160 km) spatial scales and is again likely to be due to the short pelagic larval duration found in sponges.
The Barrier analysis suggested a barrier to gene flow between adjacent Atlantic and Bay-side sites in the Middle Keys. The AMOVA confirmed that more genetic variation was found across the barrier than among sites on the same side of the barrier. In addition, the Florida-only PCoA showed the Atlantic sites (along with Waltz Key) separated from the rest of the sites by the second axis. These results suggest that dispersal through the channels between the islands of the Keys archipelago is limited, at least in the Middle Keys where we sampled. This is somewhat surprising considering the strong tidal flux through these channels (Smith 1994; Smith and Lee 2003), however, weakly-swimming larvae caught in the tidal flow may struggle to settle in areas close to the channels before being transported offshore or into the Bay. Furthermore, larval exchange could be limited by spatially and temporally variable inflow and outflow through the channels (Smith 1994; Yeung et al. 2001; Lee and Smith 2002). That being said, despite moderate statistical support for a barrier, the FST value across the barrier was only 0.033, showing low genetic differentiation. Furthermore, they did not form separate populations in Geneland and FLOCK analyses. This indicates that although genetic differentiation is higher than would be expected due to distance alone, it is only a weak barrier to gene flow. More substantial population structure was found in the seagrass Syringodium filiforme between the Bay and Atlantic sides of the Keys in the same area (Bijak et al. 2018). This is likely due to vegetative propagation of S. filiforme through the sediments compared to larval propagation of S. vesparium through the water column.
The Barrier analysis also showed a putative barrier occurring between the Middle Keys and Lower Keys sites. However, this had low bootstrap support, and is likely to be an artefact of the isolation by distance pattern in the area, rather than a physical or oceanographic barrier to dispersal (Meirmans 2012).
Other patterns of population structure within Florida did not correlate with known physical or oceanographic features. For example, sponges near Waltz Key (a semi-isolated lagoon) were genetically different than those at many other sites in the Lower Keys, but not those in the Middle Keys. Although counterintuitive, this is not uncommon. Unexpected patterns of fine-scale genetic structure have also been observed in other sponges found along the Florida Keys reef tract (DeBiasse et al. 2010; Chaves-Fonnegra et al. 2015). Furthermore, a dispersal model based on water circulation patterns and larval characteristics did not accurately predict genetic connectivity patterns for A. cervicornis across the Florida reef tract, with the genetic data revealing more complex connections than the model predicted (Drury et al. 2018). Such patterns of chaotic genetic patchiness in the marine environment can be caused by ‘sweepstakes reproductive success’, the random survival of certain larval cohorts due to oceanographic conditions (Hedgecock 1982, 1994; Hedgecock and Pudovkin 2011). These effects can be found in species with high fecundity and high larval mortality. Alternatively, variable current regimes, as found in the Florida Keys (Lee et al. 1992), can result in temporally variable dispersal pathways. Both of these situations could lead to spatially heterogeneous genetic structure through genetic drift.
Sponge restoration implications
Our results have important implications for sponge restoration practice. Genetic diversity in S. vesparium is naturally high and, in addition, clonality is low, with only two identical genotypes found in our dataset. To maintain these genetic diversity levels, restoration should be carried out though the selection of genetically-diverse donor sponges. Donor sponges should not be extensively fragmented to produce a number of genetically identical transplants in a single location; instead, minimal fragmentation of many individuals and transplantation of whole sponges should be used. By maintaining high genetic diversity, restored populations can uphold evolutionary potential and resilience against future stressors, as well as avoid the negative fitness consequences of inbreeding. As our results indicate an absence of population clusters within the Keys, strong local adaptation does not appear to be present. This indicates that outbreeding depression is not a concern, and sourcing donor sponges does not have to be restricted to certain sites or environmental conditions.
Our findings highlight the importance of restoration work in Florida. Connectivity on the regional scale was low in our study, suggesting that immigration and gene flow into Florida may be limited. Populations in Cuba or the Gulf of Mexico may be more connected to Florida that the sites sampled here. However, patterns observed in this study suggest that migration is likely to be limited due to the oceanographic distances and limited pelagic larval duration. Active management on the local scale is therefore likely to be of vital importance to ensure that population numbers are maintained.
We show that connectivity is present over the range of the Keys, and we did not observe genetically isolated sites that would need to be prioritised for restoration action. However, our results also imply that connectivity in Florida is unpredictable, as we observed unexplained fine-scale structure. Furthermore, isolation by distance suggests that dispersal is distance-limited. These results show that natural repopulation of barren areas may be slow, especially if healthy populations are moderately distant. This may be compounded by the loss of acoustic larval recruitment cues in the area, itself caused by loss of sponge-associated endosymbionts (Butler et al. 2016). Given the crucial role of this important keystone species, sponge restoration is an important strategy in facilitating a more rapid return to ecosystem function following mass mortality events. However, this approach must be coupled with thorough investigation into the causes of the ongoing mass mortalities and ecosystem instability in the Florida Keys, and the implementation of measures to mitigate these issues. Furthermore, genetic diversity and its distribution among sites should be monitored regularly to ensure that genetic variation is maintained throughout the restoration program. This can now be accomplished relatively quickly using the molecular tools described in this study.
Data accessibility
Microsatellite sequences are logged in NCBI GenBank (accession numbers KX758633- KX758644), and raw Illumina sequences of Spheciospongia vesparium genomic DNA are deposited in the NCBI Sequence Read Archive (accession reference SRP158118).
References
Alberto F (2009) MsatAllele_1.0: An R package to visualize the binning of microsatellite alleles. J Hered 100:394–397. https://doi.org/10.1093/jhered/esn110
Baums IB, Johnson ME, Devlin-Durante MK, Miller MW (2010) Host population genetic structure and zooxanthellae diversity of two reef-building coral species along the Florida Reef Tract and wider Caribbean. Coral Reefs 29:835–842. https://doi.org/10.1007/s00338-010-0645-y
Bautista-Guerrero E, Carballo JL, Maldonado M (2014) Abundance and reproductive patterns of the excavating sponge Cliona vermifera: A threat to Pacific coral reefs? Coral Reefs 33:259–266. https://doi.org/10.1007/s00338-013-1094-1
Bell JJ, Smith D, Hannan D et al (2014) Resilience to disturbance despite limited dispersal and self-recruitment in tropical barrel sponges: implications for conservation and management. PLoS ONE 9:e91635. https://doi.org/10.1371/journal.pone.0091635
Bellinger MR, Johnson JA, Toepfer J, Dunn P (2003) Loss of genetic variation in Greater Prairie Chickens following a population bottleneck in Wisconsin, U.S.A. Conserv Biol 17:717–724. https://doi.org/10.1046/j.1523-1739.2003.01581.x
Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29:1165–1188. https://doi.org/10.1214/aos/1013699998
Berry DL, Goleski JA, Koch F et al (2015) Shifts in cyanobacterial strain dominance during the onset of harmful algal blooms in Florida Bay, USA. Microb Ecol 70:361–371. https://doi.org/10.1007/s00248-014-0564-5
Bijak AL, van Dijk KJ, Waycott M (2018) Population structure and gene flow of the tropical seagrass, Syringodium filiforme, in the Florida Keys and subtropical Atlantic region. PLoS ONE 13:1–18. https://doi.org/10.1371/journal.pone.0203644
Blacket M, Robin C, Good R et al (2012) Universal primers for fluorescent labelling of PCR fragments- an efficient and cost-effective approach to genotyping by fluorescence. Mol Ecol Resour 12:456–463. https://doi.org/10.1111/j.1755-0998.2011.03104.x
Brazeau DA, Sammarco PW, Gleason DF (2005) A multi-locus genetic assignment technique to assess sources of Agaricia agaricites larvae on coral reefs. Mar Biol 147:1141–1148. https://doi.org/10.1007/s00227-005-0022-5
Bristol RM, Tucker R, Dawson DA et al (2013) Comparison of historical bottleneck effects and genetic consequences of re-introduction in a critically endangered island passerine. Mol Ecol 22:4644–4662. https://doi.org/10.1111/mec.12429
Brown RR, Davis CS, Leys SP (2017) Clones or clans: the genetic structure of a deep-sea sponge, Aphrocallistes vastus, in unique sponge reefs of British Columbia, Canada. Mol Ecol 26:1045–1059. https://doi.org/10.1111/mec.13982
Butler MJ, Dolan TW (2017) Potential impacts of everglades restoration on lobster and hard bottom communities in the Florida Keys, FL (USA). Estuaries Coasts 40:1523–1539. https://doi.org/10.1007/s12237-017-0256-8
Butler MJ, Hunt JH, Herrnkind WF et al (1995) Cascading disturbances in Florida Bay, USA: cyanobacteria blooms, sponge mortality, and implications for juvenile spiny lobsters Panulirus argus. Mar Ecol Prog Ser 129:119–125. https://doi.org/10.3354/meps129119
Butler IV MJ, Behringer DC, Valentine MM (2017) Commercial sponge fishery impacts on the population dynamics of sponges in the Florida Keys, FL(USA). Fish Res 190:113–121
Butler M, Paris C, Goldstein J et al (2011) Behavior constrains the dispersal of long-lived spiny lobster larvae. Mar Ecol Prog Ser 422:223–237. https://doi.org/10.3354/meps08878
Butler J, Stanley JA, Butler MJ (2016) Underwater soundscapes in near-shore tropical habitats and the effects of environmental degradation and habitat restoration. J Exp Mar Biol Ecol 479:89–96. https://doi.org/10.1016/j.jembe.2016.03.006
Butler MJ, Weisz JB, Butler J (2018) The effects of water quality on back-reef sponge survival and distribution in the Florida Keys, Florida (USA). J Exp Mar Biol Ecol 503:92–99. https://doi.org/10.1016/j.jembe.2018.03.001
Chandler LM, Walters LJ, Sharp WC, Hoffman EA (2017) Genetic structure of natural and broodstock populations of the long-spined sea urchin, Diadema antillarum, throughout the Florida Keys. Bull Mar Sci 93:1–9. https://doi.org/10.5343/bms.216.1101
Chapuis M-P, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631. https://doi.org/10.1093/molbev/msl191
Charlesworth D, Charlesworth D, Willis JH, Willis JH (2009) The genetics of inbreeding depression. Nat Rev Genet 10:783–796. https://doi.org/10.1038/nrg2664
Chaves-Fonnegra A, Feldheim KA, Secord J, Lopez JV (2015) Population structure and dispersal of the coral-excavating sponge Cliona delitrix. Mol Ecol 24:1447–1466. https://doi.org/10.1111/mec.13134
Chiappone M, Sullivan KM (1994) Ecological structure and dynamics of nearshore hard-bottom communities in the Florida Keys. Bull Mar Sci 54:747–756
Chybicki IJ, Burczyk J (2009) Simultaneous estimation of null alleles and inbreeding coefficients. J Hered 100:106–113. https://doi.org/10.1093/jhered/esn088
Coelho MAG, Lasker HR (2016) Larval behavior and settlement dynamics of a ubiquitous Caribbean octocoral and its implications for dispersal. Mar Ecol Prog Ser 561:109–121. https://doi.org/10.3354/meps11941
Colella MA, Ruzicka RR, Kidney JA et al (2012) Cold-water event of January 2010 results in catastrophic benthic mortality on patch reefs in the Florida Keys. Coral Reefs 31:621–632. https://doi.org/10.1007/s00338-012-0880-5
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Cowen R, Sponaugle S (2009) Larval dispersal and marine population connectivity. Ann Rev Mar Sci 1:443–466. https://doi.org/10.1146/annurev.marine.010908.163757
Cowen RK, Paris CB, Srinivasan A (2006) Scaling of connectivity in marine populations. Science 311:522–527. https://doi.org/10.1126/science.1122039
Culley TM, Stamper TI, Stokes RL et al (2013) An efficient technique for primer development and application that integrates fluorescent labeling and multiplex PCR. Appl Plant Sci 1:1300027. https://doi.org/10.3732/apps.1300027
Dailianis T, Tsigenopoulos CS, Dounas C, Voultsiadou E (2011) Genetic diversity of the imperilled bath sponge Spongia officinalis Linnaeus, 1759 across the Mediterranean Sea: patterns of population differentiation and implications for taxonomy and conservation. Mol Ecol 20:3757–3772. https://doi.org/10.1111/j.1365-294X.2011.05222.x
de Bakker DM, Meesters EHWG, van Bleijswijk JDL et al (2016) Population genetic structure, abundance, and health status of two dominant benthic species in the Saba Bank National Park, Caribbean Netherlands: Montastraea cavernosa and Xestospongia muta. PLoS ONE 11:e0155969. https://doi.org/10.1371/journal.pone.0155969
DeBiasse MB, Richards VP, Shivji MS (2010) Genetic assessment of connectivity in the common reef sponge, Callyspongia vaginalis (Demospongiae: Haplosclerida) reveals high population structure along the Florida reef tract. Coral Reefs 29:47–55. https://doi.org/10.1007/s00338-009-0554-0
DeBiasse MB, Richards VP, Shivji MS, Hellberg ME (2016) Shared phylogeographical breaks in a Caribbean coral reef sponge and its invertebrate commensals. J Biogeogr 43:2136–2146. https://doi.org/10.1111/jbi.12785
Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Ser B 39:1–38. https://doi.org/10.1111/j.2517-6161.1977.tb01600.x
Dray S, Dufour AB, Chessel D (2007) The ade4 Package - II: Two-table and K-table methods. R News 7:47–52
Drury C, Schopmeyer S, Goergen E et al (2017) Genomic patterns in Acropora cervicornis show extensive population structure and variable genetic diversity. Ecol Evol. https://doi.org/10.1002/ece3.3184
Drury C, Paris CB, Kourafalou VH, Lirman D (2018) Dispersal capacity and genetic relatedness in Acropora cervicornis on the Florida Reef Tract. Coral Reefs 37:585–596. https://doi.org/10.1007/s00338-018-1683-0
Duchesne P, Turgeon J (2012) FLOCK provides reliable solutions to the “number of populations” problem. J Hered 103:734–743. https://doi.org/10.1093/jhered/ess038
Duran S, Pascual M, Estoup A, Turon X (2004) Strong population structure in the marine sponge Crambe crambe (Poecilosclerida) as revealed by microsatellite markers. Mol Ecol 13:511–522. https://doi.org/10.1046/j.1365-294X.2004.02080.x
Ehlers A, Worm B, Reusch TBH (2008) Importance of genetic diversity in eelgrass Zostera marina for its resilience to global warming. Mar Ecol Prog Ser 355:1–7. https://doi.org/10.3354/meps07369
Ereskovsky A V. (2018) Sponge Reproduction. In: Skinner MK (ed) Encycl. Reprod., 2nd edn. pp 485–490
Evans SM, Vergés A, Poore AGB (2017) Genotypic diversity and short-term response to shading stress in a threatened seagrass: Does low diversity mean low resilience? Front Plant Sci 8:1417. https://doi.org/10.3389/fpls.2017.01417
Fiore CL, Freeman CJ, Kujawinski EB (2017) Sponge exhalent seawater contains a unique chemical profile of dissolved organic matter. PeerJ 5:e2870. https://doi.org/10.7717/peerj.2870
Fourqurean JW, Robblee MB (1999) Florida Bay: A history of recent ecological changes. Estuaries 22:345–357
Frankham R (2005) Genetics and extinction. Biol Conserv 126:131–140. https://doi.org/10.1016/j.biocon.2005.05.002
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24:2610–2618. https://doi.org/10.1111/mec.13139
Garant D, Forde SE, Hendry AP (2007) The multifarious effects of dispersal and gene flow on contemporary adaptation. Funct Ecol 21:434–443. https://doi.org/10.1111/j.1365-2435.2006.01228.x
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Gi G, Mortier F, Estoup A (2005) Geneland: a computer package for landscape genetics. Mol Ecol Notes 5:712–715. https://doi.org/10.1111/j.1471-8286.2005.01031.x
Giles EC, Saenz-Agudelo P, Hussey NE et al (2015) Exploring seascape genetics and kinship in the reef sponge Stylissa carteri in the Red Sea. Ecol Evol 5:2487–2502. https://doi.org/10.1002/ece3.1511
González-Rivero M, Ereskovsky AV, Schönberg CHL et al (2013) Life-history traits of a common Caribbean coral-excavating sponge, Cliona tenuis (Porifera: Hadromerida). J Nat Hist 47:2815–2834. https://doi.org/10.1080/00222933.2013.802042
Griffiths SM, Fox G, Briggs PJ et al (2016) A Galaxy-based bioinformatics pipeline for optimised, streamlined microsatellite development from Illumina next-generation sequencing data. Conserv Genet Resour 8:481–486. https://doi.org/10.1007/s12686-016-0570-7
Guardiola M, Frotscher J, Uriz J (2012) Genetic structure and differentiation at a short-time scale of the introduced calcarean sponge Paraleucilla magna to the western Mediterranean. Hydrobiologia 687:71–84. https://doi.org/10.1007/s10750-011-0948-1
Guardiola M, Frotscher J, Uriz M-J (2016) High genetic diversity, phenotypic plasticity, and invasive potential of a recently introduced calcareous sponge, fast spreading across the Atlanto-Mediterranean basin. Mar Biol 163:123. https://doi.org/10.1007/s00227-016-2862-6
Guillot G, Mortier F, Estoup A (2005) GENELAND: a computer package for landscape genetics. Mol Ecol Notes 5:712–715
Guillot G, Santos F, Estoup A (2008) Analysing georeferenced population genetics data with Geneland: a new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics 24:1406–1407. https://doi.org/10.1093/bioinformatics/btn136
Hedgecock D (1982) Genetic consequences of larval retention: theoretical and methodological aspects. In Kennedy V (ed) Estuarine comparisons. Academic Press, New York, pp 553–568
Hedgecock D (1994) Does variance in reproductive success limit effective population sizes of marine organisms? In Beaumont AR (ed) Genetics and evolution of aquatic organisms. Chapman & Hall, New York, pp 122–134
Hedgecock D, Pudovkin AI (2011) Sweepstakes reproductive success in highly fecund marine fish and shellfish: a review and commentary. Bull Mar Sci 87:971–1002. https://doi.org/10.5343/bms.2010.1051
Herrnkind WF, Butler MJ IV, Hunt JH, Childress M (1997) Role of physical refugia: implications from a mass sponge die-off in a lobster nursery in Florida. Mar Freshw Res 48:759. https://doi.org/10.1071/MF97193
Hicke JA, Allen CD, Desai AR et al (2012) Effects of biotic disturbances on forest carbon cycling in the United States and Canada. Glob Chang Biol 18:7–34. https://doi.org/10.1111/j.1365-2486.2011.02543.x
Hoer DR, Tommerdahl JP, Lindquist NL, Martens CS (2018) Dissolved inorganic nitrogen fluxes from common Florida Bay (U.S.A.) sponges. Limnol Oceanogr. https://doi.org/10.1002/lno.10960
Hughes AR, Stachowicz JJ (2004) Genetic diversity enhances the resistance of a seagrass ecosystem to disturbance. Proc Natl Acad Sci USA 101:8998–9002. https://doi.org/10.1073/pnas.0402642101
Jombart T (2008) Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026. https://doi.org/10.1111/j.1365-294X.2008.03887.x
Kearney KA, Butler M, Glazer R et al (2015) Quantifying Florida Bay habitat suitability for fishes and invertebrates under climate change scenarios. Environ Manage 55:836–856. https://doi.org/10.1007/s00267-014-0336-5
Keenan K, Mcginnity P, Cross TF et al (2013) DiveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol 4:782–788. https://doi.org/10.1111/2041-210X.12067
Kough AS, Paris CB (2015) The influence of spawning periodicity on population connectivity. Coral Reefs 34:753–757. https://doi.org/10.1007/s00338-015-1311-1
Kough AS, Paris CB, Behringer DC, Butler MJ IV (2015) Modelling the spread and connectivity of waterbourne marine pathogens: the case of PaV1 in the Caribbean. ICES J Mar Sci 72:i139–i146. https://doi.org/10.1093/icesjms/fsu209
Kourafalou VH, Kang H (2012) Florida current meandering and evolution of cyclonic eddies along the Florida Keys Reef Tract: are they interconnected? J Geophys Res Ocean. https://doi.org/10.1029/2011JC007383
Lauzon-Guay AJ-S, Scheibling RE (2007) Importance of spatial population characteristics on the fertilization rates of sea urchins. Biol Bull 212:195–205
Lee TN, Williams E (1999) Mean distribution and seasonal variability of coastal currents and temperature in the Florida Keys with implications for larval recruitment. Bull Mar Sci 64:35–56
Lee TN, Smith N (2002) Volume transport variability through the Florida Keys tidal channels. Cont Shelf Res 22:1361–1377. https://doi.org/10.1016/S0278-4343(02)00003-1
Lee TN, Rooth C, Williams E et al (1992) Influence of Florida Current, gyres and wind-driven circulation on transport of larvae and recruitment in the Florida Keys coral reefs. Cont Shelf Res 12:971–1002. https://doi.org/10.1016/0278-4343(92)90055-O
Levitan DR (1991) Influence of body size and population density on fertilization success and reproductive output in a free-spawning invertebrate. Biol Bull 181:261–268. https://doi.org/10.2307/1542097
López-Legentil S, Pawlik JR (2009) Genetic structure of the Caribbean giant barrel sponge Xestospongia muta using the I3–M11 partition of COI. Coral Reefs 28:157–165. https://doi.org/10.1007/s00338-008-0430-3
Maldonado M (2006) The ecology of the sponge larva. Can J Zool 84:175–194. https://doi.org/10.1139/z05-177
Maldonado M, Riesgo A (2008) Reproduction in Porifera: a synoptic overview. Treballs la SCB 59:29–49
Manni F, Guérard E, Heyer E (2004) Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier’s algorithm. Hum Biol 76:173–190
Mariani S, Uriz MJ, Turon X (2000) Larval bloom of the oviparous sponge Cliona viridis: Coupling of larval abundance and adult distribution. Mar Biol 137:783–790. https://doi.org/10.1007/s002270000400
Mariani S, Piscitelli MP, Uriz MJ (2001) Temporal and spatial co-occurrence in spawning and larval release of Cliona viridis (Porifera: Hadromerida). J Mar Biol Assoc United Kingdom 81:565–567. https://doi.org/10.1017/S0025315401004246
Meirmans PG (2012) The trouble with isolation by distance. Mol Ecol 21:2839–2846. https://doi.org/10.1111/j.1365-294X.2012.05578.x
Meirmans PG, Van Tiendener PH (2004) Genotype and Genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794. https://doi.org/10.1111/j.1471-8286.2004.00770.x
Monmonier MS (1973) Maximum-difference barriers: an alternative numerical regionalization method. Geogr Anal 5:245–261. https://doi.org/10.1111/j.1538-4632.1973.tb01011.x
Muhling BA, Smith RH, Vásquez-Yeomans L et al (2013) Larval fish assemblages and mesoscale oceanographic structure along the Mesoamerican Barrier Reef System. Fish Oceanogr 22:409–428. https://doi.org/10.1111/fog.12031
Narum SR (2006) Beyond Bonferroni: Less conservative analyses for conservation genetics. Conserv Genet 7:783–787. https://doi.org/10.1007/s10592-005-9056-y
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Padua A, Cunha HA, Klautau M (2017) Gene flow and differentiation in a native calcareous sponge (Porifera) with unknown dispersal phase. Mar Biodivers 1–11: https://doi.org/10.1007/s12526-017-0742-z
Paetkau D, Slade R, Burden M, Estoup A (2004) Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol Ecol 13:55–65
Pante E, Simon-Bouhet B (2013) marmap: A package for importing, plotting and analyzing bathymetric and topographic data in R. PLoS ONE 8:e73051. https://doi.org/10.1371/journal.pone.0073051
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research- an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Pérez-Portela R, Noyer C, Becerro MA (2015) Genetic structure and diversity of the endangered bath sponge Spongia lamella. Aquat Conserv Mar Freshw Ecosyst 25:365–379. https://doi.org/10.1002/aqc.2423
Peterson B, Chester C, Jochem F, Fourqurean JW (2006) Potential role of sponge communities in controlling phytoplankton blooms in Florida Bay. Mar Ecol Prog Ser 328:93–103
Piry S, Alapetite A, Cornuet J-M et al (2004) GENECLASS2: A software for genetic assignment and first-generation migrant detection. J Hered 95:536–539. https://doi.org/10.1093/JHERED/ESH074
Piscitelli M, Corriero G, Gaino E, Uriz MJ (2011) Reproductive cycles of the sympatric excavating sponges Cliona celata and Cliona viridis in the Mediterranean Sea. Invertebr Biol 130:1–10. https://doi.org/10.1111/j.1744-7410.2010.00216.x
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci USA 94:9197–9201. https://doi.org/10.1073/pnas.94.17.9197
Raymond M, Rousset F (1995) GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenicism. J Hered 86:248–249
R Core Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.Rproject.org/
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237. https://doi.org/10.1046/j.1523-1739.2003.01236.x
Richards VP, Bernard AM, Feldheim KA, Shivji MS (2016) Patterns of population structure and dispersal in the long-lived “redwood” of the coral reef, the giant barrel sponge (Xestospongia muta). Coral Reefs 35:1097–1107. https://doi.org/10.1007/s00338-016-1435-y
Riesgo A, Pérez-Portela R, Pita L et al (2016) Population structure and connectivity in the Mediterranean sponge Ircinia fasciculata are affected by mass mortalities and hybridization. Heredity (Edinb) 117:427–439. https://doi.org/10.1038/hdy.2016.41
Riesgo A, Taboada S, Pérez-Portela R et al (2019) Genetic diversity, connectivity and gene flow along the distribution of the emblematic Atlanto-Mediterranean sponge Petrosia ficiformis (Haplosclerida, Demospongiae). BMC Evol Biol 19:1–18. https://doi.org/10.1186/s12862-018-1343-6
Rosell D, Uriz MJ (2002) Excavating and endolithic sponge species (Porifera) from the Mediterranean: Species descriptions and identification key. Org Divers Evol 2:55–86. https://doi.org/10.1078/1439-6092-00033
Rosenberg NA (2004) DISTRUCT: A program for the graphical display of population structure. Mol Ecol Notes 4:137–138. https://doi.org/10.1046/j.1471-8286.2003.00566.x
Rousset F (2008) Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Saenz-Agudelo P, Jones GP, Thorrold SR, Planes S (2011) Connectivity dominates larval replenishment in a coastal reef fish metapopulation. Proc R Soc B Biol Sci 278:2954–2961. https://doi.org/10.1098/rspb.2010.2780
Sbordoni V, De Matthaeis E, Cobolli Sbordoni M et al (1986) Bottleneck effects and the depression of genetic variability in hatchery stocks of Penaeus japonicus (Crustacea, Decapoda). Aquaculture 57:239–251. https://doi.org/10.1016/0044-8486(86)90202-4
Schönberg CHL (2002) Pione lampa, a bioeroding sponge in a worm reef. Hydrobiologia 482:49–68. https://doi.org/10.1023/A:1021258901538
Selkoe K, Toonen R (2011) Marine connectivity: a new look at pelagic larval duration and genetic metrics of dispersal. Mar Ecol Prog Ser 436:291–305. https://doi.org/10.3354/meps09238
Shafer AB, Wolf JB, Alves PC et al (2015) Genomics and the challenging translation into conservation practice. Trends Ecol Evol 30:78–87
Smith NP (1994) Long-term Gulf to Atlantic transport through tidal channels in the Florida Keys. Bull Mar Sci 54:602–609
Smith NP, Lee TN (2003) Volume transport through tidal channels in the Middle Florida Keys. J Coast Res 19:254–260
Sorte CJB, Davidson VE, Franklin MC et al (2017) Long-term declines in an intertidal foundation species parallel shifts in community composition. Glob Chang Biol 23:341–352. https://doi.org/10.1111/gcb.13425
Sponaugle S, Lee T, Kourafalou V, Pinkard D (2005) Florida Current frontal eddies and the settlement of coral reef fishes. Limnol Oceanogr 50:1033–1048. https://doi.org/10.4319/lo.2005.50.4.1033
Stevely JM, Sweat DE, Bert TM, et al (2010) Sponge mortality at Marathon and Long Key, Florida: patterns of species response and population recovery. In Proceedings of the 63rd Gulf and Caribbean Fisheries Institute, vol 63, pp 384–400
Sweeney BW, Bott TL, Jackson JK et al (2004) Riparian deforestation, stream narrowing, and loss of stream ecosystem services. Proc Natl Acad Sci USA 101:14132–14137. https://doi.org/10.1073/pnas.0405895101
Szpiech ZA, Jakobsson M, Rosenberg NA (2008) ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24:2498–2504. https://doi.org/10.1093/bioinformatics/btn478
Taboada S, Riesgo A, Wiklund H et al (2018) Implications of population connectivity studies for the design of marine protected areas in the deep sea: An example of a demosponge from the Clarion-Clipperton Zone. Mol Ecol. https://doi.org/10.1111/mec.14888
Taylor HR, Dussex N, van Heezik Y (2017) Bridging the conservation genetics gap by identifying barriers to implementation for conservation practitioners. Glob Ecol Conserv 10:231–242
Tellier M-AS, Bertelsen R (2008) Monitoring the flora and fauna of the nearshore hardbottom habitats of the Florida Keys. F2196-05-08-F Final Report. Florida Fish and Wildlife Conservation Commission, Fish and Wildlife Research Institute, Marathon, FL
Thomson JA, Burkholder DA, Heithaus MR et al (2015) Extreme temperatures, foundation species, and abrupt ecosystem change: an example from an iconic seagrass ecosystem. Glob Chang Biol 21:1463–1474. https://doi.org/10.1111/gcb.12694
Trakhtenbrotl A, Nathan R, Perry G, Richardson DM (2005) The importance of long-distance dispersal in biodiversity conservation. Divers Distrib 11:173–181
Truelove NK, Kough AS, Behringer DC et al (2017) Biophysical connectivity explains population genetic structure in a highly dispersive marine species. Coral Reefs 36:233–244. https://doi.org/10.1007/s00338-016-1516-y
Valentine MM, Butler MJ (2019) Sponges structure water-column characteristics in shallow tropical coastal ecosystems. Mar Ecol Prog Ser 608:133–147. https://doi.org/10.3354/meps12758
van der Meer MH, Horne JB, Gardner MG, et al (2013) Limited contemporary gene flow and high self-replenishment drives peripheral isolation in an endemic coral reef fish. Ecol Evol. https://doi.org/10.1002/ece3.584
Wall CC, Rodgers BS, Gobler CJ, Peterson BJ (2012) Responses of loggerhead sponges Spechiospongia vesparium during harmful cyanobacterial blooms in a sub-tropical lagoon. Mar Ecol Prog Ser 451:31–43. https://doi.org/10.3354/meps09537
Warburton FE (1966) The behaviour of sponge larvae. Ecology 47:672–674
Whitlock MC (2000) Fixation of new alleles and the extinction of small populations: drift load, beneficial alleles and sexual selection. Evolution (N Y) 54:1855. https://doi.org/10.1554/0014-3820(2000)054[1855:FONAAT]2.0.CO;2
Wright S (1943) Isolation by distance. Genetics 28:114–138
Wright S (1949) The genetical structure of populations. Ann Eugen 15:323–354. https://doi.org/10.1111/j.1469-1809.1949.tb02451.x
Wulff J (2006) Rapid diversity and abundance decline in a Caribbean coral reef sponge community. Biol Conserv 127:167–176. https://doi.org/10.1016/j.biocon.2005.08.007
Xavier JR, Van SRWM, Breeuwer JAJ, Menken SBJ (2010) Phylogeography, genetic diversity and structure of the poecilosclerid sponge Phorbas fictitius at oceanic islands. Contrib to Zool 79:119–129
Yeung C, Jones DL, Criales MM et al (2001) Influence of coastal eddies and counter-currents on the influx of spiny lobster, Panulirus argus, postlarvae into Florida Bay. Mar Freshw Res 52:1217–1232
Acknowledgements
Many thanks for assistance in sampling to T. Pérez, C. Freeman, F. Clever, L. Gittens, the Forest and Marine Reserves Association of Caye Caulker (FAMRACC), the PACOTILLES campaign team and crew of the RV Antea, and the Old Dominion University and University of Florida 2014 field teams. Thanks to P. Briggs at the University of Manchester Bioinformatics Core Facility, and to N. Truelove for sequencing assistance. Many thanks to T. Pérez for providing helpful comments and discussion on the manuscript. This work was funded by a PhD studentship awarded to S. Griffiths by the Natural Environment Research Council (NERC). Funding for the field collections in Florida was provided by a Nature Conservancy-NOAA Community-based Habitat Restoration Grant (GMT-ODU-091512) to M. Butler and D. Behringer. Fieldwork in Barbuda was funded by ship time awarded to T. Pérez and C. Fauvelot by Flotte Océanographique Française. Field collection of sponge tissue in the Florida Keys was conducted under permits issued to M. Butler by the Everglades National Park (EVER-00401), the Florida Keys National Marine Sanctuary (FKNMS-2014–042), and the Florida Fish and Wildlife Conservation Commission (SAL-13-0582A-SR). Research collections in The Bahamas were conducted under a permit granted by The Bahamas Department of Marine Resources to S. Griffiths and R. Preziosi (MAMR/FIS/17). Sampling in Belize was conducted under a permit granted by Belize Fisheries Department to S. Griffiths and R. Preziosi (0.00023–13). Samples in Barbuda were collected under permits issued to T. Pérez and C. Fauvelot as part of the PACOTILLES campaign (https://dx.doi.org/10.17600/15005300; https://dx.doi.org/10.17600/15005200).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by David Hawksworth.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article belongs to the Topical Collection: Coastal and marine biodiversity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Griffiths, S.M., Taylor-Cox, E.D., Behringer, D.C. et al. Using genetics to inform restoration and predict resilience in declining populations of a keystone marine sponge. Biodivers Conserv 29, 1383–1410 (2020). https://doi.org/10.1007/s10531-020-01941-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-020-01941-7