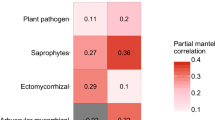
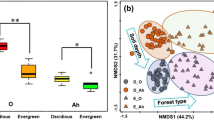
Abstract
Here we characterize and compare the diversity of belowground fungal communities of maples (Sapindaceae: Acer) varying in both nativity and weediness, and interpret our findings in the context of multiple non-exclusive theories on tree invasions and fungal associations. We made our fungal community comparisons based on high-throughput Illumina sequencing of the internal transcribed spacer region of fungal ribosomal DNA of soil samples associated with the roots of different species of maple collected from six sites throughout Central Pennsylvania. In our system, we found that weedy species, regardless of nativity, had the greatest soil fungal richness and that the nonnative invasive Norway maple had the highest abundance of mycorrhizal mutualists. Despite that much of the fungal community variability in our system was attributable to inter-site variability, we found that the core fungal communities associated with nonnative tree species were an inclusively larger set that included nearly all of those associated with native trees in addition to many not found with the natives, and the core communities of non-weedy species were largely a subset of those associated with weedy maples. In addition to confirming the strong influence that site variation has on soil fungal communities, our findings are also largely consistent with positive feedback from native fungal communities, possible co-invasion by fungal associates that are only associated with the nonnative trees, and generally add to the growing number of studies that have observed a greater abundance of mutualists associated with invasive trees that interact with arbuscular mycorrhizal fungi.





Similar content being viewed by others
References
Alexander HD, Arthur MA (2010) Implications of a predicted shift from upland oaks to red maple on forest hydrology and nutrient availability. Can J For Res 40:716–726. doi:10.1139/X10-029
Bainard LD, Klironomos JN, Gordon AM (2011) The mycorrhizal status and colonization of 26 tree species growing in urban and rural environments. Mycorrhiza 21:91–96. doi:10.1007/s00572-010-0314-6
Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H (2010) ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 10:189. doi:10.1186/1471-2180-10-189
Bogar LM, Dickie IA, Kennedy PG (2015) Testing the co-invasion hypothesis: ectomycorrhizal fungal communities on Alnus glutinosa and Salix fragilis in New Zealand. Divers Distrib 21:268–278. doi:10.1111/ddi.12304
Bunn RA, Lekberg Y, Gallagher C, Rosendahl S, Ramsey PW (2014) Grassland invaders and their mycorrhizal symbionts: a study across climate and invasion gradients. Ecol Evol 4:794–805. doi:10.1002/ece3.917
Bunn RA, Ramsey PW, Lekberg Y (2015) Do native and invasive plants differ in their interactions with arbuscular mycorrhizal fungi? A meta-analysis. J Ecol 103:1547–1556. doi:10.1111/1365-2745.12456
Burgess TI, Crous CJ, Slippers B, Hantula J, Wingfield MJ (2016) Tree invasions and biosecurity: eco-evolutionary dynamics of hitchhiking fungi. AoB Plants 8:plw076. doi:10.1093/aobpla/plw076
Callaway RM, Thelen GC, Rodriguez A, Holben WE (2004) Soil biota and exotic plant invasion. Nature 427:731–733. doi:10.1038/nature02322
Callaway RM, Cippolini D, Barto K, Thelen GC (2008) Novel weapons: invasive plant suppresses fungal mutualists in America but not in its native Europe. Ecology 89:1043–1055. doi:10.1890/07-0370.1
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi:10.1038/nmeth.f.303
Coats VC, Rumpho ME (2014) The rhizosphere microbiota of plant invaders: an overview of recent advances in the microbiomics of invasive plants. Front Microbiol 5:368. doi:10.3389/fmicb.2014.00368
Dawkins K, Esiobu N (2017) Arbuscular and ectomycorrhizal fungi associated with the invasive Brazilian Pepper Tree (Schinus terebinthifolius) and two native plants in South Florida. Front Microbiol 8:665. doi:10.3389/fmicb.2017.00665
Dawson W, Schrama M (2016) Identifying the role of soil microbes in plant invasions. J Ecol 104:1211–1218. doi:10.1111/1365-2745.12619
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi:10.1093/bioinformatics/btq461
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi:10.1038/nmeth.2604
Eppinga MB, Rietkerk M, Dekker SC, De Ruiter PC, Van der Putten WH (2006) Accumulation of local pathogens: a new hypothesis to explain exotic plant invasions. Oikos 114:168–176. doi:10.1111/j.2006.0030-1299.14625.x
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. doi:10.1111/j.1365-294X.1993.tb00005.x
Gornish ES, Fierer N, Barberán A (2016) Associations between an invasive plant (Taeniatherum caput-medusae, Medusahead) and soil microbial communities. PLoS ONE 11:e0163930. doi:10.1371/journal.pone.0163930
Hakamata T, Yamamoto S (2008) Growth and development of broad leaved tree seedlings treated with VA mycorrhizal materials and Japanese cedar-charcoal. Bulletin of the Shizuoka Research Institute of Agriculture and Forestry (Japan), pp 79–85
Hayward J, Horton TR, Nuñez MA (2015a) Ectomycorrhizal fungal communities coinvading with Pinaceae host plants in Argentina: Gringos bajo el bosque. New Phytol 208:497–506. doi:10.1111/nph.13453
Hayward J, Horton T, Pauchard A, Nunez M (2015b) A single ectomycorrhizal fungal species can enable a Pinus invasion. Ecology 96:1438–1444. doi:10.1890/14-1100.1
Horsley SB, Long RP, Bailey SW, Hallett RA, Wargo PM (2002) Health of eastern North American sugar maple forests and factors affecting decline. North J Appl For 19:34–44
Hynson NA, Merckx VS, Perry BA, Treseder KK (2013) Identities and distributions of the co-invading ectomycorrhizal fungal symbionts of exotic pines in the Hawaiian Islands. Biol Invasions 15:2373–2385. doi:10.1007/s10530-013-0458-3
Ibáñez I, McCarthy-Neumann S (2015) Effects of mycorrhizal fungi on tree seedling growth: quantifying the parasitism–mutualism transition along a light gradient. Can J For Res 46:48–57. doi:10.1139/cjfr-2015-0327
Klironomos JN (2002) Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417:67–70. doi:10.1038/417067a
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H (2013) Towards a unified paradigm for sequence-based identification of Fungi. Mol Ecol 22:5271–5277. doi:10.1111/mec.12481
McGuire KL, Payne SG, Palmer MI, Gillikin CM, Keefe D, Kim SJ, Gedallovich SM, Discenza J, Rangamannar R, Koshner JA, Massmann AL, Orazi G, Essene A, Leff JW, Fierer N (2013) Digging the New York City skyline: soil fungal communities in green roofs and city parks. PLoS ONE 8:e58020. doi:10.1371/journal.pone.0058020
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. doi:10.1371/journal.pone.0061217
Meinhardt KA, Gehring CA (2012) Disrupting mycorrhizal mutualisms: a potential mechanism by which exotic tamarisk outcompetes native cottonwoods. Ecol Appl 22:532–549. doi:10.1890/11-1247.1
Moora M, Berger S, Davidson J, Opik M, Bommarco R, Bruelheide H, Kuhn I, Kunin WE, Metsis M, Rortais A, Vanatoa A, Stout JC, Truusa M, Westphal C, Zobel M, Walther GR (2011) Alien plants associate with widespread generalist arbuscular mycorrhizal fungal taxa: evidence from a continental-scale study using massively parallel 454 sequencing. J Biogeogr 38:1305–1317. doi:10.1111/j.1365-2699.2011.02478.x
Muth NZ, Pigliucci M (2006) Traits of invasives reconsidered: phenotypic comparisons of introduced invasive and introduced noninvasive plant species within two closely related clades. Am J Bot 93:188–196. doi:10.3732/ajb.93.2.188
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248. doi:10.1016/j.funeco.2015.06.006
Nijjer S, Rogers WE, Lee CTA, Siemann E (2008) The effects of soil biota and fertilization on the success of Sapium sebiferum. Appl Soil Ecol 38:1–11. doi:10.1016/j.apsoil.2007.08.002
Nuñez MA, Dickie IA (2014) Invasive belowground mutualists of woody plants. Biol Invasions 16:645–661. doi:10.1007/s10530-013-0612-y
Oliveros JC (2007-2015) Venny. An interactive tool for comparing lists with Venn’s diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html
Paulson JN, Stine OC, Bravo HC, Pop M (2013) Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10:1200–1202. doi:10.1038/nmeth.2658
Reinhart KO, Callaway RM (2004) Soil biota facilitate exotic Acer invasions in Europe and North America. Ecol Appl 14:1737–1745. doi:10.1890/03-5204
Rhoads AF, Block TA (2005) Trees of Pennsylvania: a complete reference guide. University of Pennsylvania Press, Philadelphia
Rodrigues RR, Pineda RP, Barney JN, Nilsen ET, Barrett JE, Williams MA (2015) Plant invasions associated with change in root-zone microbial community structure and diversity. PLoS ONE 10:e0141424. doi:10.1371/journal.pone.0141424
Stinson KA, Campbell SA, Powell JR, Wolfe BE, Callaway RM, Thelen GC, Hallett SG, Prati D, Klironomos J (2006) Invasive plant suppresses the growth of native tree seedlings by disrupting belowground mutualisms. PLoS Biol 4:e140. doi:10.1371/journal.pbio.0040140
Tonn N (2016) The effect of urbanization of the mycorrhizal associations and survival of three species of eastern hardwoods. Dissertation, University of Michigan
Trexler R, Solomon C, Brislawn CJ, Wright JR, Rosenberger A, McClure EE, Grube AM, Peterson MP, Keddache M, Mason OU, Hazen TC, Grant CJ, Lamendella R (2014) Assessing impacts of unconventional natural gas extraction on microbial communities in headwater stream ecosystems in Northwestern Pennsylvania. Front Microbiol 5:1–13. doi:10.3389/fmicb.2014.00522
Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer, New York
Wiseman PE, Wells C (2005) Soil inoculum potential and arbuscular mycorrhizal colonization of Acer rubrum in forested and developed landscapes. J Arboric 31:296–302
Yang Q, Wei S, Shang L, Carrillo J, Gabler CA, Nijjer S, Li B, Siemann E (2015) Mycorrhizal associations of an invasive tree are enhanced by both genetic and environmental mechanisms. Ecography 38:1112–1118. doi:10.1111/ecog.00965
Acknowledgements
Funding for this project was provided by GCAT-SEEK through the National Science Foundation (DBI-1248096), by Howard Hughes Medical Institute, and by Juniata College Student Research Fellowship Committee. Special thanks to Maple Brook Nursery and Fox Hill Gardens for their nursery stock to be sampled, Jeff Meadows for access to the Juniata College and Peace Chapel sampling locations, and the US Army Corps of Engineers for access to the Raystown Lake and Seven Points Marina sampling locations. Additional thanks for analytical assistance from Caleb Madder and William Bernard. Thanks to Vince Buonaccorsi for help with the experimental design.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Richness estimates calculated for fungal communities present in soil samples collected from the roots of maples varying in nativity and invasiveness (n = 46). Estimates were calculated from the untransformed OTU table after the data were filtered to remove samples with less than 1000 sequence counts from the analysis and rarefied down to the lowest sequencing depth across all samples. In each plot, alpha diversity values were plotted along the y-axis while the maple species were plotted along the x-axis. Richness estimates were made using the metrics for Observed OTUs (left), Chao1 (middle), and inverse of Simpson’s Diversity Index (right). Error bars were created to show standard error within a 95% confidence interval of each Chao1 estimate. Plots and estimates were generated with the Phyloseq R-package (McMurdie and Holmes 2013) along with a visual overlay produced by the ggplot2 R-package (Wickham 2009) (TIFF 250 kb)
Supplementary Fig. 2
Richness estimates calculated for fungal communities present in soil samples collected from the roots of maples varying in nativity and invasiveness (n = 46). Estimates were calculated from the untransformed OTU table after the data were filtered to remove samples with less than 1000 sequence counts from the analysis and rarefied down to the lowest sequencing depth across all samples. In each plot, alpha diversity values were plotted along the y-axis while the maple species were plotted along the x-axis. Richness estimates were made using the metric for Observed OTUs. Richness plots were facetted by sample site. Plots and estimates were generated with the Phyloseq R-package (McMurdie and Holmes 2013) along with a visual overlay produced by the ggplot2 R-package (Wickham 2009) (TIFF 285 kb)
Supplementary Fig. 3
Non-metric multidimensional scaling (NMDS) plots generated for soil samples collected from the roots of maples varying in nativity and invasiveness (n = 46). Plots were made from an OTU table normalized using metagenomeSeq’s Cumulative Sum Scaling algorithm after the data were filtered to remove samples with less than 1000 sequence counts from the analysis and trimmed to not retain singleton reads. Sample points were plotted on several axes of variation and represented in 2D space by the axes showing the most variation between samples. Samples were colored by (a) maple species and (b) site location. Plots and estimates were generated with the Phyloseq R-package (McMurdie and Holmes 2013) along with a visual overlay produced by the ggplot2 R-package (Wickham 2009) (TIFF 655 kb)
Rights and permissions
About this article
Cite this article
Toole, D.R., Cannon, G.H., Brislawn, C.J. et al. Differences in soil fungal assemblages associated with native and non-native tree species of varying weediness. Biol Invasions 20, 891–904 (2018). https://doi.org/10.1007/s10530-017-1580-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-017-1580-4