Abstract
Objective
To develop an efficient synthetic promoter library for fine-tuned expression of target genes in Corynebacterium glutamicum.
Results
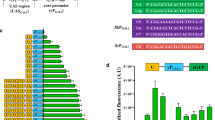
A synthetic promoter library for C. glutamicum was developed based on conserved sequences of the − 10 and − 35 regions. The synthetic promoter library covered a wide range of strengths, ranging from 1 to 193% of the tac promoter. 68 promoters were selected and sequenced for correlation analysis between promoter sequence and strength with a statistical model. A new promoter library was further reconstructed with improved promoter strength and coverage based on the results of correlation analysis. Tandem promoter P70 was finally constructed with increased strength by 121% over the tac promoter. The promoter library developed in this study showed a great potential for applications in metabolic engineering and synthetic biology for the optimization of metabolic networks.
Conclusions
To the best of our knowledge, this is the first reconstruction of synthetic promoter library based on statistical analysis of C. glutamicum.





Similar content being viewed by others
References
Blazeck J, Alper HS (2013) Promoter engineering: recent advances in controlling transcription at the most fundamental level. Biotechnol J 8:46–58
Chung S-C, Park J-S, Yun J, Park JH (2017) Improvement of succinate production by release of end-product inhibition in Corynebacterium glutamicum. Metab Eng 40:157–164
De Mey M, Maertens J, Lequeux GJ, Soetaert WK, Vandamme EJ (2007) Construction and model-based analysis of a promoter library for E. coli: an indispensable tool for metabolic engineering. BMC Biotechnol 7:34
Jajesniak P, Seng Wong T (2015) From genetic circuits to industrial-scale biomanufacturing: bacterial promoters as a cornerstone of biotechnology. AIMS Bioeng 2:277–296
Jensen K, Alper H, Fischer C, Stephanopoulos G (2006) Identifying functionally important mutations from phenotypically diverse sequence data. Appl Environ Microbiol 72:3696–3701
Kim MJ, Yim SS, Choi JW, Jeong KJ (2016) Development of a potential stationary-phase specific gene expression system by engineering of SigB-dependent cg3141 promoter in Corynebacterium glutamicum. Appl Microbiol Biotechnol 100:4473–4483
Kubota T, Watanabe A, Suda M, Kogure T, Hiraga K, Inui M (2016) Production of para-aminobenzoate by genetically engineered Corynebacterium glutamicum and non-biological formation of an N-glucosyl byproduct. Metab Eng 38:322–330
Liu X et al (2016) Expression of recombinant protein using Corynebacterium Glutamicum: progress, challenges and applications. Crit Rev Biotechnol 36:652–664
Meng H, Wang Y (2015) Cis-acting regulatory elements: from random screening to quantitative design. Quant Biol 3:107–114
Patek M, Nesvera J, Guyonvarch A, Reyes O, Leblon G (2003) Promoters of Corynebacterium glutamicum. J Biotechnol 104:311–323
Ravasi P et al (2015) High-level production of Bacillus cereus phospholipase C in Corynebacterium glutamicum. J Biotechnol 216:142–148
Redden H, Alper HS (2015) The development and characterization of synthetic minimal yeast promoters. Nat Commun 6:7810
Rhodius VA, Mutalik VK, Gross CA (2012) Predicting the strength of UP-elements and full-length E-coli sigma(E) promoters. Nucl Acids Res 40:2907–2924
Rytter JV, Helmark S, Chen J, Lezyk MJ, Solem C, Jensen PR (2014) Synthetic promoter libraries for Corynebacterium glutamicum. Appl Microbiol Biotechnol 98:2617–2623
Shang X et al (2018) Native promoters of Corynebacterium glutamicum and its application in l-lysine production. Biotechnol Lett 40:383–391
Wu G, Yan Q, Jones JA, Tang YJ, Fong SS, Koffas MAG (2016) Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol 34:652–664
Xu J, Han M, Zhang J, Guo Y, Zhang W (2014) Metabolic engineering Corynebacterium glutamicum for the l-lysine production by increasing the flux into l-lysine biosynthetic pathway. Amino Acids 46:2165–2175
Yim SS, An SJ, Kang M, Lee J, Jeong KJ (2013) Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol Bioeng 110:2959–2969
Zhang B, Ren LQ, Yu M, Zhou Y, Ye BC (2017a) Enhanced l-ornithine production by systematic manipulation of l-ornithine metabolism in engineered Corynebacterium glutamicum S9114. Bioresour Technol 250:60–68
Zhang W, Zhao Z, Yang Y, Liu X, Bai Z (2017b) Construction of an expression vector that uses the aph promoter for protein expression in Corynebacterium glutamicum. Plasmid 94:1–6
Acknowledgements
This work was supported by National Natural Science Foundation of China (NSFC-21576200, NSFC-21776209, NSFC-21621004 and NSFC-21390201) and Natural Science Foundation of Tianjin (No. 15JCQNJC06000).
Supporting information
Supplementary Table 1—Sequence of the individual promoters.
Author information
Authors and Affiliations
Corresponding author
Additional information
Shuanghong Zhang and Dingyu Liu are the first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, S., Liu, D., Mao, Z. et al. Model-based reconstruction of synthetic promoter library in Corynebacterium glutamicum. Biotechnol Lett 40, 819–827 (2018). https://doi.org/10.1007/s10529-018-2539-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-018-2539-y