Abstract
Objective
To identify useful native promoters of Corynebacterium glutamicum for fine-tuning of gene expression in metabolic engineering.
Results
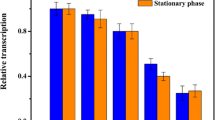
Sixteen native promoters of C. glutamicum were characterized. These promoters covered a strength range of 31-fold with small increments and exhibited relatively stable activity during the whole growth phase using β-galactosidase as the reporter. The mRNA level and enzymatic activity of the lacZ reporter gene exhibited high correlation (R 2 = 0.96) under the control of these promoters. Sequence analysis found that strong promoters had high similarity of the -10 hexamer to the consensus sequence and preference of the AT-rich UP element upstream the -35 region. To test the utility of the promoter library, the characterized native promoters were applied to modulate the sucCD-encoded succinyl-CoA synthetase expression for l-lysine overproduction.
Conclusions
The native promoters with various strengths realize the efficient and precise regulation of gene expression in metabolic engineering of C. glutamicum.




Similar content being viewed by others
References
Albersmeier A, Pfeifer-Sancar K, Rückert C, Kalinowski J (2017) Genome-wide determination of transcription start sites reveals new insights into promoter structures in the actinomycete Corynebacterium glutamicum. J Biotechnol 257:99–109
Becker J, Wittmann C (2015) Advanced biotechnology: metabolically engineered cells for the bio-based production of chemicals and fuels, materials, and health-care products. Angew Chem Int Ed 54:3328–3350
Becker J, Zelder O, Häfner S, Schröder H, Wittmann C (2011) From zero to hero—Design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metab Eng 13:159–168
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Han SO, Inui M, Yukawa H (2007) Expression of Corynebacterium glutamicum glycolytic genes varies with carbon source and growth phase. Microbiology 153:2190–2202
Kind S, Becker J, Wittmann C (2013) Increased lysine production by flux coupling of the tricarboxylic acid cycle and the lysine biosynthetic pathway—Metabolic engineering of the availability of succinyl-CoA in Corynebacterium glutamicum. Metab Eng 15:184–195
Lee J (2014) Development and characterization of expression vectors for Corynebacterium glutamicum. J Microbiol Biotechnol 24:70–79
Liu Q, Ouyang SP, Kim J, Chen GQ (2007) The impact of PHB accumulation on l-glutamate production by recombinant Corynebacterium glutamicum. J Biotechnol 132:273–279
Mentz A, Neshat A, Pfeifer-Sancar K, Pühler A, Rückert C, Kalinowski J (2013) Comprehensive discovery and characterization of small RNAs in Corynebacterium glutamicum ATCC 13032. BMC Genom 14:714
Nešvera J, Pátek M (2011) Tools for genetic manipulations in Corynebacterium glutamicum and their applications. Appl Microbiol Biotechnol 90:1641–1654
Ooyen JV, Noack S, Bott M, Reth A, Eggeling L (2012) Improved l-lysine production with Corynebacterium glutamicum and systemic insight into citrate synthase flux and activity. Biotechnol Bioeng 109:2070–2081
Park JU, Jo JH, Kim YJ, Chung SS, Lee JH, Lee HH (2008) Construction of heat-inducible expression vector of Corynebacterium glutamicum and C. ammoniagenes: fusion of lambda operator with promoters isolated from C. ammoniagenes. J Microbiol Biotechnol 18:639–647
Pátek M, Nešvera J (2011) Sigma factors and promoters in Corynebacterium glutamicum. J Biotechnol 154:101–113
Pátek M, Nešvera J, Guyonvarch A, Reyes O (2003) Promoters of Corynebacterium glutamicum. J Biotechnol 104:311–323
Rytter JV, Helmark S, Chen J, Lezyk MJ, Solem C, Jensen PR (2014) Synthetic promoter libraries for Corynebacterium glutamicum. Appl Microbiol Biotechnol 98:2617–2623
Schweitzer JE, Stolz M, Diesveld R, Etterich H, Eggeling L (2009) The serine hydroxymethyltransferase gene glyA in Corynebacterium glutamicum is controlled by GlyR. J Biotechnol 139:214–221
Tanaka Y, Ehira S, Teramoto H, Inui M, Yukawa H (2012) Coordinated regulation of gnd, which encodes 6-phosphogluconate dehydrogenase, by the two transcriptional regulators GntR1 and RamA in Corynebacterium glutamicum. J Bacteriol 194:6527–6536
Vidal-Aroca F, Giannattasio M, Brunelli E, Vezzoli A, Plevani P, Muzi-Falconi M, Bertoni G (2006) One-step high-throughput assay for quantitative detection of β-galactosidase activity in intact gram-negative bacteria, yeast, and mammalian cells. Biotechniques 40:433–440
Yim SS, An SJ, Kang M, Lee J, Jeong KJ (2013) Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol Bioeng 110:2959–2971
Zhang Y, Shang XL, Lai SJ, Zhang GQ, Liang Y, Wen TY (2012) Development and application of an arabinose-inducible expression system by facilitating inducer uptake in Corynebacterium glutamicum. Appl Environ Microbiol 78:5831–5838
Zhang Y, Cai JY, Shang XL, Wang B, Liu SW, Chai X, Tan TW, Zhang Y, Wen TY (2017) A new genome-scale metabolic model of Corynebacterium glutamicum and its application. Biotechnol Biofuels 10:169
Zhao ZH, Liu XX, Zhang W, Yang YK, Dai XF, Bai ZH (2016) Construction of genetic parts from the Corynebacterium glutamicum genome with high expression activities. Biotechnol Lett 38:2119–2126
Acknowledgements
This work was supported by grants from National Hi-Tech Research and Development Program of China (2014AA021203), the Science and Technology Service Network Initiative of the Chinese Academy of Sciences (KFJ-EW-STS-078) and National Natural Science Foundation of China (31100074).
Supporting information
Supplementary Table 1—Strains and plasmids used.
Supplementary Table 2—Primers used.
Supplementary Fig. 1—Sequence analysis of the sixteen promoters.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human and animal participants
The research performed did not involve human participants and/or animals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shang, X., Chai, X., Lu, X. et al. Native promoters of Corynebacterium glutamicum and its application in l-lysine production. Biotechnol Lett 40, 383–391 (2018). https://doi.org/10.1007/s10529-017-2479-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-017-2479-y