Abstract
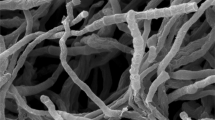
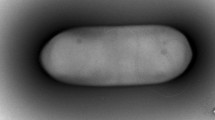
Strain H2R21T, a novel actinobacterium, isolated from a forest soil sample collected from Heybeliada, Istanbul, Turkey, and a polyphasic approach was used for characterisation of the strain. Chemotaxonomic and morphological characterisation of strain H2R21T indicated that it belongs to the genus Nonomuraea. 16S rRNA gene sequence similarity showed that the strain is closely related to Nonomuraea purpurea 1SM4-01T (99.1%) and Nonomuraea solani CGMCC 4.7037T (98.4%). DNA–DNA relatedness values were found to be lower than 70% between the isolate and its phylogenetic neighbours N. purpurea 1SM4-01T, N. solani CGMCC 4.7037T and Nonomuraea rhizophila YIM 67092T. The whole cell hydrolysates of strain H2R21T were found to contain meso-diaminopimelic acid as the diagnostic diamino acid and glucose, madurose, mannose and ribose as the cell sugars. The polar lipids were identified as phosphatidylglycerol, diphosphatidylglycerol, phosphatidylmethylethanolamine, phosphatidylethanolamine, hydroxy-phosphatidylethanolamine, dihydroxy-phosphatidylethanolamine, phosphatidylinositol, glycophosphatidylinositol, two glycophospholipids and two unidentified lipids. The predominant menaquinones were identified as MK-9(H4) and MK-9(H6). The major fatty acids were found to be iso-C16:0, iso-C16:0 2OH and C17:0 10-methyl. On the basis of DNA–DNA relatedness data and some phenotypic characteristics, it is evident that strain H2R21T can be distinguished from the closely related species in the genus Nonomuraea. Thus, it is concluded that strain H2R21T represents a novel species of the genus Nonomuraea, for which the name Nonomuraea insulae sp. nov. is proposed. The type strain is H2R21T (= DSM 102915T = CGMCC 4.7338T = KCTC 39769T).


Similar content being viewed by others
References
Chun J, Goodfellow M (1995) A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Bacteriol 45:240–245
Collins MD (1985) Isoprenoid quinone analysis in bacteria classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 267–287
Dalmastri C, Gastaldo L, Marcone GL, Binda E, Congiu T, Marinelli F (2016) Classification of Nonomuraea sp. ATCC 39727, an actinomycete that produces the glycopeptide antibiotic A40926, as Nonomuraea gerenzanensis sp. nov. Int J Syst Evol Microbiol 66:912–921
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:143–153
Fang BZ, Hua ZS, Han MX, Zhang ZT, Wang YH, Yang ZW, Zhang WQ, Xiao M, Li WJ (2017) Nonomuraea cavernae sp. nov., a novel actinobacterium isolated from a karst cave sample. Int J Syst Evol Microbiol 67:4692–4697
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogeny: an approach using the bootstrap. Evolution 39:783–791
Hayakawa M, Nonomura H (1987) Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J Ferment Technol 65:501–509
Huss VAR, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridisation from renaturation rates. Syst Appl Microbiol 4:184–192
Jones KL (1949) Fresh isolates of actinomycetes in which the presence of sporogenous aerial mycelia is a fluctuating characteristic. J Bacteriol 57:141–145
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Kämpfer P (2012) Genus VI. Nonomuraea. In: Whitman WB, Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Ludwig W, Suzuki KI, Parte A (eds) Bergey’s manual of systematic bacteriology. The Actinobacteria, vol 5, 2nd edn. Springer, New York, pp 1844–1861
Kämpfer P, Kroppenstedt RM (1996) Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can J Microbiol 42:989–1005
Kämpfer P, Kroppenstedt RM, Grün-Wollny L (2005) Nonomuraea kuesteri sp. nov. Int J Syst Evol Microbiol 55:847–851
Kelly KL (1964) Inter-society color council-national bureau of standards color-name charts illustrated with centroid colors. US Government Printing Office, Washington, DC
Kluge AG, Farris FS (1969) Quantitative phyletics and the evolution of anurans. Syst Zool 18:1–32
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger. J Liq Chromatogr 5:2359–2387
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lechevalier MP, Lechevalier HA (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol 20:435–443
Li J, Zhao GZ, Huang HY, Zhu WY, Lee JC, Xu LH, Kim CJ, Li WJ (2011) Nonomuraea endophytica sp. nov., an endophytic actinomycete isolated from Artemisia annua L. Int J Syst Evol Microbiol 61:757–761
Li Z, Song W, Zhao JW, Zhuang XX, Zhao Y, Xiang WS, Wang XJ (2017) Nonomuraea glycinis sp. nov., a novel actinomycete isolated from the root of Black soya bean [Glycine max (L.) Merr]. Int J Syst Evol Microbiol 67:5026–5031
Mandel M, Marmur J (1968) Use of ultraviolet absorbance temperature profile for determining the guanine plus cytosine content of DNA. Methods Enzymol 12B:195–206
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Nakaew N, Sungthong R, Yokota A, Lumyong S (2012) Nonomuraea monospora sp. nov., an actinomycete isolated from cave soil in Thailand, and emended description of the genus Nonomuraea. Int J Syst Evol Microbiol 62:3007–3012
Nash P, Krent MM (1991) Culture media. In: Balows A, Hauser WJ, Herrmann KL, Isenberg HD, Shadomy HJ (eds) Manual of clinical microbiology, 3rd edn. American Society for Microbiology, Washington, DC, pp 1268–1270
Niemhom N, Chutrakul C, Suriyachadkun C, Thawai C (2017) Nonomuraea stahlianthi sp. nov., an endophytic actinomycete isolated from the stem of Stahlianthus campanulatus. Int J Syst Evol Microbiol 67:2879–2884
Rachniyom H, Matsumoto A, Indananda C, Duangmal K, Takahashi Y, Thamchaipenet A (2015) Nonomuraea syzygii sp. nov., an endophytic actinomycete isolated from the roots of a jambolan plum tree (Syzygium cumini L. Skeels). Int J Syst Evol Microbiol 65:1234–1240
Saitou N, Nei M (1987) The neighbor-joining method. A new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. Technical Note 101. MIDI Inc: Newark
Shen Y, Jia FY, Liu CX, Li JS, Guo SY, Zhou SY, Wang XJ, Xiang WS (2016) Nonomuraea zeae sp. nov., isolated from the rhizosphere of corn (Zea mays L.). Int J Syst Evol Microbiol 66:2259–2264
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Sripreechasak P, Phongsopitanun W, Supong K, Pittayakhajonwut P, Kudo T, Ohkuma M, Tanasupawat S (2017) Nonomuraea rhodomycinica sp. nov., isolated from peat swamp forest soil in Thailand. Int J Syst Evol Microbiol 67:1683–1687
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Suksaard P, Mingma R, Srisuk N, Matsumoto A, Takahashi Y, Duangmal K (2016) Nonomuraea purpurea sp. nov., an actinomycete isolated from mangrove sediment. Int J Syst Evol Microbiol 66:4987–4992
Sungthong R, Nakaew N (2015) The genus Nonomuraea: a review of a rare actinomycete taxon for novel metabolites. J Basic Microbiol 55:554–565
Waksman SA (1961) The actinomycetes, classification, identification and description of genera and species, vol 2. Williams & Wilkins, Baltimore
Waksman SA (1967) The actinomycetes. A summary of current knowledge. Ronald Press, New York
Wang XJ, Zhao JW, Liu CX, Wang JD, Shen Y, Jia FY, Wang L, Zhang J, Yu C, Xiang WS (2013) Nonomuraea solani sp. nov., a novel actinomycete isolated from eggplant root (Solanum melongena L.). Int J Syst Evol Microbiol 63:2418–2423
Wang S, Liu C, Zhang Y, Zhao J, Zhang X, Yang L, Wang X, Xiang W (2014) Nonomuraea guangzhouensis sp. nov., and Nonomuraea harbinensis sp. nov., two novel actinomycetes isolated from soil. Antonie Van Leeuwenhoek 105:109–118
Wang F, Shi JD, Huang YJ, Wu YY, Deng XM (2017) Nonomuraea ceibae sp. nov., a new actinobacterium isolated from Ceiba speciosa rhizosphere. Int J Syst Evol Microbiol 67:1158–1162
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneath PHA, Sackin MJ (1983) Numerical classification of Streptomyces and related genera. J Gen Microbiol 129:1743–1813
Xi L, Zhang L, Ruan J, Huang Y (2011) Nonomuraea maritima sp. nov., isolated from coastal sediment. Int J Syst Evol Microbiol 61:2740–2744
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhang Z, Wang Y, Ruan J (1998) Reclassification of Thermomonospora and Microtetraspora. Int J Syst Bacteriol 48:411–422
Zhao GZ, Li J, Huang HY, Zhu WY, Xu LH, Li WJ (2011) Nonomuraea rhizophila sp. nov., an actinomycete isolated from rhizosphere soil. Int J Syst Evol Microbiol 61:2141–2145
Acknowledgements
This research was supported by Ondokuz Mayis University (OMU), project no. PYO.FEN.1904.13.004. We gratefully acknowledges Dr. Kannika Duangmal from Kasetsart University in Thailand for help to us in obtaining one of the type species.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saricaoglu, S., Nouioui, I., Ay, H. et al. Nonomuraea insulae sp. nov., isolated from forest soil. Antonie van Leeuwenhoek 111, 2051–2059 (2018). https://doi.org/10.1007/s10482-018-1097-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-018-1097-6