Abstract
Objective
Intravoxel incoherent motion (IVIM) shows great potential in many applications, e.g., tumor tissue characterization. To reduce image-quality demands, various IVIM analysis approaches restricted to the diffusion coefficient (D) and the perfusion fraction (f) are increasingly being employed. In this work, the impact of estimation approach for D and f is studied.
Materials and methods
Four approaches for estimating D and f were studied: segmented IVIM fitting, least-squares fitting of a simplified IVIM model (sIVIM), and Bayesian fitting of the sIVIM model using marginal posterior modes or posterior means. The estimation approaches were evaluated in terms of bias and variability as well as ability for differentiation between tumor and healthy liver tissue using simulated and in vivo data.
Results
All estimation approaches had similar variability and ability for differentiation and negligible bias, except for the Bayesian posterior mean of f, which was substantially biased. Combined use of D and f improved tumor-to-liver tissue differentiation compared with using D or f separately.
Discussion
The similar performance between estimation approaches renders the segmented one preferable due to lower numerical complexity and shorter computational time. Superior tissue differentiation when combining D and f suggests complementary biologically relevant information.
Similar content being viewed by others
Introduction
Diffusion and perfusion magnetic resonance imaging (MRI) are frequently used for in vivo tumor tissue characterization [1, 2]. Studies have shown that both techniques can have a predictive role in therapy response assessment, including a potential to indicate treatment effects earlier than standard morphological evaluation [3]. An improved ability to differentiate between various malignant brain tumors has been shown for combined use of diffusion and perfusion MRI [4]. In liver metastases from neuroendocrine tumors (NETs), both techniques have been shown to reflect changes induced by therapy [5].
Both diffusion and perfusion are motion of water molecules on a subvoxel scale. The intravoxel incoherent motion (IVIM) model aims to describe the effect of these two motions on the signal intensity in diffusion-weighted images [6]. Successful estimation of the IVIM model parameters would thus provide both diffusion and perfusion information noninvasively from a single imaging sequence. Since diffusion weighting sensitizes the image to motion, perfusion-related IVIM parameters contain information related to the amount of flowing blood in the capillaries and its velocity. On the other hand, dynamic contrast-enhanced (DCE) MRI, used in most studies on tumor microvasculature, also provides information on the permeability and surface area of microvessels [7]. Still, promising results on tumor tissue characterization based on IVIM have been shown that warrant further studies [8,9,10].
The IVIM model is commonly formulated as a two-compartment model, as follows:
where S(b) is the signal at a diffusion weighting with b value b, S0 is the signal without diffusion weighting, f is the perfusion fraction, D is the diffusion coefficient, and D* is the pseudodiffusion coefficient [6]. Due to the higher rate of motion of water molecules in blood, D* is usually expected to be at least one order of magnitude greater than D.
Because of the characteristics of the IVIM model and typical values of its parameters, estimating D* has proved to be difficult, demanding high-quality data and, as a result, long examination times [11]. Therefore, estimating only D and f has been employed in several recent studies [12,13,14]. This still enables extraction of both diffusion (D) and perfusion (f) information while reducing the demands on image quality in terms of signal-to-noise ratio (SNR).
Multiple studies concerning approaches for fitting the IVIM model have been conducted, e.g., [16, 17, 20]. However, evaluation of approaches for estimating D and f only is, to our knowledge, limited to a subanalysis in a single study [16]. Furthermore, while Bayesian estimation techniques have shown great promise for the full IVIM model, evaluation of the potential improvement in estimating D and f only is lacking [21,22,23]. When the performance of a set of estimators is compared, general measures such as the bias and variability of the estimates can be used. However, while small bias and variability are desired features, the clinical usefulness of the estimated parameters should also be taken into account [24]. This could, e.g., include the ability to differentiate between tissue types, such as tumor and normal tissue.
The aim of this study was to investigate the impact of the estimation approach for the IVIM model restricted to the parameters D and f. Specifically, effects on estimation bias and variability as well as ability to differentiate between NET liver metastases and healthy liver tissue were studied.
Materials and methods
Parameter estimation
Two major approaches for estimating only D and f have been proposed: one is based on a specialized model-fitting procedure, and one uses a special case of the IVIM model (Eq. 1), both assuming b values in certain ranges and that D* ≫ D. In the former approach, often referred to as segmented fitting, estimation is done in two steps [15,16,17]. In the first step, data from b values below a certain threshold (bthr) are omitted. If bthr is large enough, the signal from the perfusion compartment is considered to be of negligible size and the IVIM model simplifies to a monoexponential model:
In the second step, f is estimated as f = 1 − A/S(0), where S(0) is the measured signal at b = 0. In the latter approach, a simplified version of the IVIM model (sIVIM) is considered:
where \(\delta \left( b \right)\) is the discrete delta function, i.e., \(\delta \left( {b = 0} \right) = 1\) and \(\delta \left( {b \ne 0} \right) = 0\) [16, 18, 19]. The model is valid for the b values b = 0 and b ≥ bthr.
Four specific approaches for estimating D and f were considered in this study:
-
1.
Segmented fitting, where D is estimated from b values ≥ 120 s/mm2 (i.e., bthr = 120 s/mm2) and f from the intercept A (Eq. 2), as described above
-
2.
Least-squares fitting of the sIVIM model (Eq. 3)
-
3.
Bayesian fitting of the sIVIM model using the marginal posterior modes
-
4.
Bayesian fitting of the sIVIM model using the posterior means.
Segmented fitting was performed using a custom-made MATLAB function with nonlinear least-squares fitting of D. Least-squares fitting of the sIVIM model was done with the MATLAB function fit with default arguments.
The Bayesian model fitting based on Eq. 3 was performed using a previously published MATLAB function for Bayesian IVIM model fittingFootnote 1 [21], which was adapted to the sIVIM model. The implementation uses a Markov chain Monte Carlo setup to sample the posterior parameter distribution from which the marginal posterior mode or posterior mean was estimated. Uniform prior distributions were used for all parameters.
For all four estimation approaches, the parameter estimates of D, f, and S0 were constrained to the ranges [0 5] µm2/ms, [0 1] and [0 2 Smax], respectively, where Smax is the maximum measured or simulated signal value depending on the context. For the segmented model fit, the constraint on f was applied by setting negative estimates to zero. For the Bayesian methods, the constraints were applied by setting the prior distributions to zero outside the specified ranges. Additional detailed information about the parameter estimation approaches can be found in the supplementary information.
Patients
MR imaging data from patients with liver metastases from small-intestine NET was obtained from a previously published randomized clinical trial of embolization methods [25]. Patients were randomly assigned to either hepatic artery embolization or radioembolization treatment and were examined with MRI before and one and 3 months after treatment. Among the 11 patients in the previous study, one failed to undergo the MR examination due to cardiac pacemaker, and one was examined with a different MR protocol, resulting in nine patients for analysis. For detailed descriptions of inclusion criteria and treatments, the reader is referred to the previous paper [25]. The MR examinations included in the study we report here were those performed before (baseline) and 3 months after treatment.
MR imaging
Respiratory-triggered diffusion weighted images (DWIs) of the upper abdomen were acquired on a Philips Achieva dStream 3T with software release 5.1.7 (Best, The Netherlands) using a single-shot spin-echo echoplanar imaging (SE-EPI) sequence with five b values (0, 120, 350, 575, 800 s/mm2, Δ ≈ 26 ms, δ ≈ 16 ms). For b > 0, three orthogonal diffusion-encoding directions were acquired. Number of directions × number of signal averages = total number of measurements at each b value, were 1 × 6 = 6, 3 × 3 = 9, 3 × 3 = 9, 3 × 6 = 18 and, 3 × 6 = 18, respectively. Other imaging parameters were: TE = 54 ms, TR = 2600 ms, half scan = 0.70, acquisition pixel size = 3 × 3 mm2, reconstructed pixel size = 1.8 × 1.8 mm2, slice thickness = 6 mm, and slice gap = 0.6 mm. Phase encoding was performed in the anterior–posterior direction, with sensitivity-encoding (SENSE) factor = 2 and a resulting bandwidth of 13.7 Hz/mm. Regions of interest (ROIs) were produced by manual delineation of the tumor border using the DWI with b = 0. ROIs were also drawn in healthy liver and spleen in the same images, avoiding large vessels. ROIs in liver and spleen were drawn such that their size was similar to the overall average tumor size. This strategy was employed to get approximately the same number of voxels for each tissue type. SNR was calculated as the signal in the image with b = 0 divided by the standard deviation of the noise, taking into account the effects of averaging. The noise level was estimated from the residuals of a monoexponential fit of data with b > 0. The resulting median SNR estimates in tumor, liver, and spleen were 16, 20, and 18, respectively.
Simulations
Simulated data were generated from the sIVIM model (Eq. 3) for the same b values and total number of measurements at each b value as for the in vivo acquisitions at three SNR levels: 10, 20, and 40. At each level 10,000 data series with Rician noise were generated based on values of D and f randomly drawn from uniform distributions with bounds [0.5, 1.5] µm2/ms and [0, 0.3], respectively. SNR refers to the measurement at b = 0 after averaging. The noise level after averaging was thus lower at the higher b values due to the larger number of averages.
Statistical analysis
The quality of parameter estimates obtained from the different estimation approaches using simulated data were compared in terms of bias and variability. This was done by studying the quantiles of the distribution of differences between estimated and simulated parameter value. For parameter estimates based on in vivo data, where the true parameter values are unknown, the relative bias and variability were studied by comparison of results between estimation approaches. To evaluate whether the b-value threshold was sufficiently high, D and f were estimated excluding data with b = 120 s/mm2 and compared with estimates based on all b values.
The ability of different estimation approaches to differentiate between tumor and healthy liver tissue was studied by constructing a classifier for each approach separately based on kernel density estimation. The performance of classifiers was quantified using a leave-subject-out cross-validation where the classifier was trained on data from all but one patient. Data from that patient was then used for testing. The training and testing procedure was repeated such that all patients were used for testing once. The classifier was trained on voxel parameter data from all patients such that for each tissue type, the tissue-specific probability density function (pdf) was estimated using the MATLAB function ksdensity with a Gaussian-shaped kernel and default arguments. Classification was performed on all voxel data from the test patient by identifying the tissue type with the highest probability based on the estimated tissue-specific pdf. The analysis was performed both in one dimension for D and f separately and in two dimensions with D and f combined. The proportion of correct classifications was averaged across the repetitions to calculate the overall performance of the classifier. The classification was performed for each time-point separately. MATLAB 2016b (MathWorks, Natick, USA) was used for all calculations and visualization.
Results
Simulations
All estimation approaches in general showed similar performance with negligible average bias, except for f based on Bayesian estimation of the posterior mean, which was positively biased (Fig. 1). By studying the dependence of quantiles on simulated values, it is apparent that the bias was exclusive to small true values of f (Fig. 2 for SNR = 20, and Supplementary Figs. S1 and S2 for SNR = 10 and 40, respectively). In Fig. 2 one can also observe that the bias of D from the same estimation approach depends strongly on the true value of f. Note that this is hidden in Fig. 1, which only shows the average bias. A similar but weaker trend was found for D when estimated with the Bayesian marginal posterior mode. No considerable differences in variability between estimation approaches were seen.
Estimation error (estimated minus true parameter value) for f (a) and D (b) based on simulated data at different signal-to-noise-ratio (SNR) levels for all four evaluated approaches: SEG segmented fitting, LSQ least-squares fitting, BMO Bayesian fitting using the posterior marginal modes, BME Bayesian fitting using the posterior means. Whiskers indicate the 1st and 99th percentiles. The horizontal black line shows zero error
In vivo
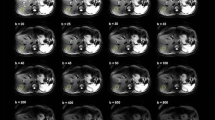
Tumors were clearly visible on the b = 0 image, enabling manual delineation without the use of information from other images. A b = 0 image from an example patient is shown in Fig. 3, along with parameter maps of D and f over a region including the delineated tumor for all estimation approaches.
The b = 0 image and corresponding intravoxel incoherent motion (IVIM) parameter maps for an example patient. Manual delineation of the tumor of interest is marked with a red dashed line in the image and a black dotted line in the parameter maps. The image region shown in the parameter maps is depicted by a red rectangle in the b = 0 image. Note that maps of a particular parameter are almost indistinguishable when compared visually
Results based on in vivo data showed similar trends as those from simulations when comparing estimation approaches, although somewhat less pronounced. The bias of f based on Bayesian estimation of the posterior mean was apparent for tissues with low perfusion fractions (tumor and spleen) but not in the liver, which in general showed higher f values. The bias manifested as a larger value of the lower quartile in Fig. 4 and is more clearly visible in Fig. 5, where parameter estimates from the segmented approach are compared with those from the other estimation approaches in a similar way as for simulated data in Fig. 2. The more subtle bias trends seen for D in Fig. 2 are, however, not manifested in Fig. 5. The difference in D and f estimates obtained with or without excluding b = 120 s/mm2 was of negligible size (median difference −0.04/ −0.05/0.001 µm2/ms and 0/0.019/0, respectively, for tumor/liver/spleen).
Estimated values of f (a) and D (b) in different tissues for the evaluated approaches: SEG segmented fitting, LSQ least-squares fitting, BMO Bayesian fitting using the posterior marginal modes, BME Bayesian fitting using the posterior means. Whiskers show the 1st and 99th percentiles. Note the elevated lower quartile of f from BME seen in both tumor and spleen and compare with Fig. 1a
Comparison of parameter estimates from in vivo data between each model-fitting approach and the segmented approach plotted as a function of parameter estimates from the segmented approach. Each plot shows the 1st, 25th, 50th, 75th, and 99th percentiles. Note that the y-axis when comparing f estimates has a different range than that in Fig. 2. The x-axis for D is also different compared with Fig. 2 due to the narrower range of estimated values of D in vivo
All tissue types displayed distinctly different distributions of D and f (Fig. 4). The variability of D was substantially smaller in healthy liver and spleen than in tumor tissue, indicating highly heterogeneous tumor tissue. Tumor and spleen tissue showed similar distribution of f. In scatter plots showing D vs. f, tumor and healthy liver tissue displays distinctly different patterns (Fig. 6). At baseline, tumor and healthy liver were observed to be more separated in the scatter plot than in separate histograms of D or f (Fig. 6). The same trend could be seen in the classification analysis where the combined use of D and f resulted in a substantially higher proportion of voxels correctly classified as tumor tissue (Fig. 7). The ability to differentiate between tissue types was similar for all evaluated estimation approaches (Figs. 6, 7). At 3 months after treatment, the tissue-specific joint distributions of D and f were substantially less structured, especially for tumor tissue (Supplementary Fig. S3), which resulted in a reduced ability to differentiate between tissue types compared with baseline (Supplementary Fig. S4). Still, the results were comparable among all evaluated estimation approaches.
Distribution of voxel values of D and f in tumor and healthy liver tissue at baseline for each model-fitting approach. Kernel density estimates of tumor (red) and liver (green) are overlaid on the scatter plots and histograms (solid line) and reproduced in the other tissue type (dashed line) for comparison. The two-dimensional kernel density estimate is represented by a contour at an arbitrarily chosen level (same in all plots)
Classification performance between tumor and healthy liver tissue for all four evaluated approaches: SEG segmented fitting, LSQ least-squares fitting, BMO Bayesian fitting using the posterior marginal modes, BME Bayesian fitting using the posterior means based on D and f at baseline. Bars show the average fraction of correctly classified voxels given by cross-validation. The classifier was trained using D, f, or both D and f (indicated under each corresponding group of bars in the graph)
Discussion
The segmented approach has been studied extensively both as part of evaluations of estimation approaches for the full IVIM model (e.g. [15, 16, 26]) and more recently for estimation limited to D and f [27, 28]. However, comparison of estimation approaches for D and f only is, to our knowledge, limited to a single study in which the segmented approach was compared with least-squares fitting of the sIVIM model [16]. That comparison was part of a larger evaluation of estimation approaches for DWI data and their applicability in prostate cancer. The current study extends beyond the previous one by applying simulations over a range of values of D and by including Bayesian approaches in the comparison. It complements the previous study by analyzing other in vivo tissue types.
The major difference between using the segmented approach and estimation based on the sIVIM model is how the parameter constraints are applied. In the segmented approach, the estimate of D is only affected by its own constraint, meaning that A > S(0) may occur. On the other hand, estimates of D obtained from fitting the sIVIM model are affected by constraints on both D and f, resulting in an implicit constraint of A ≤ S(0), which resulted in a negative bias on D for small values of f where the constraint has a large impact. No such trend could be seen for the segmented approach. Estimates of f are affected by constraints on both D and f regardless of estimation approach. The similar results for all estimation approaches regarding f is therefore expected (Fig. 2, Supplementary Figs. S1, S2, and Fig. 5).
The results based on simulated and in vivo data agreed in the sense that no major differences on parameter estimates could be seen between estimation approaches, except for the bias of f based on Bayesian estimation of the posterior mean. Some of the bias trends seen in the simulation were not distinguishable in the results from in vivo data. This could in part be due to the somewhat different ranges and combinations of D and f in the simulations and in vivo data, where from a parameter-estimation perspective, least favorable combinations of D and f potentially were missing or less frequently represented in the in vivo data. Still, the overall trend from both simulations and in vivo evaluation is that only minor differences in bias and variability can be seen between estimation approaches, which is in concordance with previous findings [16]. The same trend is seen for the ability to differentiate between tumor and healthy liver tissue, which was similar for all estimation approaches, including the substantially biased Bayesian posterior mean of f.
Since none of the studied estimation approaches was superior regarding bias/variability or tissue differentiation, the numerical complexity and computational speed may be included as factors when choosing a preferred approach. In such case, the segmented approach is highly preferable, since it reduces the estimation to a one-dimensional optimization problem for estimating D and a simple calculation of f. This is in contrast with when the sIVIM model is used, for which multiple parameters must be estimated simultaneously, resulting in substantially increased numerical complexity and computational time.
Choice of prior distribution for Bayesian IVIM model fitting has previously been shown to have a substantial effect on parameter estimates from the full IVIM model [21]. Due to the exclusion of D* in the sIVIM model, the model is considerably less flexible and therefore is likely less susceptible to noise. Choice of prior distribution should thus be less influential when using the sIVIM model unless highly informative priors are employed. Uniform priors were chosen in this study due to their simplicity and lack of subjectivity. Still, more informative priors may provide more robust parameter estimation if the assumptions incorporated in the priors are suitable. Data-driven informative priors have been proposed for the full IVIM model with promising results [22, 29], but rigorous validation needs to be done to avoid errors due to inappropriate assumptions [30].
Combined use of D and f improved the ability to differentiate between NET metastases and healthy liver tissue compared with the use of D or f alone, suggesting that complementary biologically relevant information is provided by the two parameters. This is in line with previous findings regarding tumor tissue characterization and therapy-response assessment [8, 13], although contradicting results have been reported for prostate cancer [16]. The distribution of D and f was substantially altered by the treatments, suggesting a therapeutic effect on at least a subset of the studied tumors. However, due to the involvement of two different treatments in this relatively small patient group and a possible range of responses, no conclusions regarding treatment-specific response assessment using IVIM parameters can be drawn.
Only one set of b values was considered in this study. Consensus regarding b values for estimating IVIM D and f has not yet been established and thus varies between studies [12, 13], although some work has been done regarding optimization of three-b-value protocols [27, 28]. However, as long as the chosen model is valid for the particular b values, choice of b values should only have minor effects on the observed estimation trends. In our study, the signal contribution from the perfusion compartment is assumed to be of negligible size at b = 120 s/mm2. The validity of this assumption depends on a sufficiently large value of D* for the particular tissue type. While both liver and spleen are associated with large values of D* [11], thereby validating the chosen b-value threshold, IVIM studies of NET liver metastases are lacking. However, the separate analysis, which excluded b = 120 s/mm2, indicated only a very small bias for D. The effects of a b-value threshold that is potentially too low should thus be negligible for the chosen b-value scheme. Nevertheless, further studies regarding optimal sets of b values should be conducted for increased comparability between studies. To adhere to the in vivo data in which the contribution from the perfusion compartment was negligible at b ≥ 120 s/mm2, simulations were performed based on the sIVIM model (Eq. 3) rather than the IVIM model (Eq. 1) with some specific value of D*. This also enabled analysis of the bias related only to the estimation approach and not to the influence of a potential contribution from the perfusion compartment. Another possible limitation of this study is the manual tumor delineation, which may have included some nontumor voxels on tumor borders. Such voxels could potentially bias the classification analysis and decrease the maximum attainable classification performance. However, the number of such voxels is likely small due to the high contrast between tumor and healthy liver in the images used for delineation. The effect on classification results should thus be negligible.
In conclusion, all evaluated estimation approaches showed similar performance, although the Bayesian posterior mean of f was substantially more biased for small true values of f. Taking numerical complexity and computational time into account, the segmented approach is preferable.
Notes
References
Padhani AR, Liu G, Mu-Koh D et al (2009) Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia 11:102–125
Padhani AR, Miles KA (2010) Multiparametric imaging of tumor response to therapy. Radiology 256:348–364
Harry VN, Semple SI, Parkin DE, Gilbert FJ (2010) Use of new imaging techniques to predict tumour response to therapy. Lancet Oncol 11:92–102
Calli C, Kitis O, Yunten N, Yurtseven T, Islekel S, Akalin T (2006) Perfusion and diffusion MR imaging in enhancing malignant cerebral tumors. Eur J Radiol 58:394–403
Liapi E, Geschwind J-F, Vossen JA, Buijs M, Georgiades CS, Bluemke DA, Kamel IR (2008) Functional MRI evaluation of tumor response in patients with neuroendocrine hepatic metastasis treated with transcatheter arterial chemoembolization. AJR Am J Roentgenol 190:67–73
Le Bihan D, Breton E, Lallemand D, Aubin ML, Vignaud J, Laval-Jeantet M (1988) Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology 168:497–505
O’Connor JPB, Jackson A, Parker GJM, Jayson GC (2007) DCE–MRI biomarkers in the clinical evaluation of antiangiogenic and vascular disrupting agents. Br J Cancer 96:189–195
Cho GY, Moy L, Kim SG, Baete SH, Moccaldi M, Babb JS, Sodickson DK, Sigmund EE (2016) Evaluation of breast cancer using intravoxel incoherent motion (IVIM) histogram analysis: comparison with malignant status, histological subtype, and molecular prognostic factors. Eur Radiol 26:2547–2558
Wu H, Zhang S, Liang C, Liu H, Liu Y, Mei Y, Liu H, Liu Z, Xu F (2017) Intravoxel incoherent motion MRI for the differentiation of benign, intermediate, and malignant solid soft-tissue tumors. J Magn Reson Imaging 46:1611–1618
Woo S, Lee JM, Yoon JH, Joo I, Han JK, Choi BI (2014) Intravoxel incoherent motion diffusion-weighted MR imaging of hepatocellular carcinoma: correlation with enhancement degree and histologic grade. Radiology 270:758–767
Lemke A, Stieltjes B, Schad LR, Laun FB (2011) Toward an optimal distribution of b values for intravoxel incoherent motion imaging. Magn Reson Imaging 29:766–776
Yuan Q, Costa DN, Sénégas J, Xi Y, Wiethoff AJ, Rofsky NM, Roehrborn C, Lenkinski RE, Pedrosa I (2017) Quantitative diffusion-weighted imaging and dynamic contrast-enhanced characterization of the index lesion with multiparametric MRI in prostate cancer patients. J Magn Reson Imaging 45:908–916
Mürtz P, Penner A-H, Pfeiffer A-K et al (2016) Intravoxel incoherent motion model—based analysis of diffusion-weighted magnetic resonance imaging with 3 b-values for response assessment in locoregional therapy of hepatocellular carcinoma. Onco Targets Ther 9:6425–6433
Penner A-H, Sprinkart AM, Kukuk GM, Gütgemann I, Gieseke J, Schild HH, Willinek WA, Mürtz P (2013) Intravoxel incoherent motion model-based liver lesion characterisation from three b-value diffusion-weighted MRI. Eur Radiol 23:2773–2783
Pekar J, Moonen CTW, van Zijl PCM (1992) On the precision of diffusion/perfusion imaging by gradient sensitization. Magn Reson Med 23:122–129
Merisaari H, Movahedi P, Perez IM et al (2017) Fitting methods for intravoxel incoherent motion imaging of prostate cancer on region of interest level: repeatability and Gleason score prediction. Magn Reson Med 77:1249–1264
Barbieri S, Donati OF, Froehlich JM, Thoeny HC (2016) Impact of the calculation algorithm on biexponential fitting of diffusion-weighted MRI in upper abdominal organs. Magn Reson Med 75:2175–2184
Sénégas J, Perkins TG, Keupp J, Stehning C, Herigault G, Smith-Miloff M, Hussain SM (2012) Towards organ-specific b values for the IVIM-based quantification of ADC: in vivo evaluation in the liver. In: Proceedings of the 20th annual meeting of ISMRM. Melbourne, Australia, p 1891
Yuan Q, Costa DN, Sénégas J, Xi Y, Wiethoff AJ, Lenkinski RE, Pedrosa I (2015) A simplified intravoxel incoherent motion model for diffusion weighted imaging in prostate cancer evaluation: comparison with monoexponential and biexponential models. In: Proceedings of the 23rd annual meeting of ISMRM. Toronto, Canada, p 3019
Cho GY, Moy L, Zhang JL et al (2015) Comparison of fitting methods and b-value sampling strategies for intravoxel incoherent motion in breast cancer. Magn Reson Med 74:1077–1085
Gustafsson O, Montelius M, Starck G, Ljungberg M (2018) Impact of prior distributions and central tendency measures on Bayesian intravoxel incoherent motion model fitting. Magn Reson Med 79:1674–1683
Orton MR, Collins DJ, Koh D-M, Leach MO (2014) Improved intravoxel incoherent motion analysis of diffusion weighted imaging by data driven Bayesian modeling. Magn Reson Med 71:411–420
Neil JJ, Bretthorst GL (1993) On the use of Bayesian probability theory for analysis of exponential decay data: an example taken from intravoxel incoherent motion experiments. Magn Reson Med 29:642–647
Jambor I, Merisaari H, Taimen P, Boström P, Minn H, Pesola M, Aronen HJ (2015) Evaluation of different mathematical models for diffusion-weighted imaging of normal prostate and prostate cancer using high b-values: a repeatability study. Magn Reson Med 73:1988–1998
Elf A-K, Andersson M, Henrikson O, Jalnefjord O, Ljungberg M, Svensson J, Wängberg B, Johanson V (2018) Radioembolization versus bland embolization for hepatic metastases from small intestinal neuroendocrine tumors: short-term results of a randomized clinical trial. World J Surg 42:506–513
Park HJ, Sung YS, Lee SS, Lee Y, Cheong H, Kim YJ, Lee MG (2017) Intravoxel incoherent motion diffusion-weighted MRI of the abdomen: the effect of fitting algorithms on the accuracy and reliability of the parameters. J Magn Reson Imaging 45:1637–1647
Meeus EM, Novak J, Dehghani H, Peet AC (2017) Rapid measurement of intravoxel incoherent motion (IVIM) derived perfusion fraction for clinical magnetic resonance imaging. Magn Reson Mater Phy. https://doi.org/10.1007/s10334-017-0656-6
While PT, Teruel JR, Vidić I, Bathen TF, Goa PE (2017) Relative enhanced diffusivity: noise sensitivity, protocol optimization, and the relation to intravoxel incoherent motion. Magn Reson Mater Phy. https://doi.org/10.1007/s10334-017-0660-x
Freiman M, Perez-Rossello JM, Callahan MJ, Voss SD, Ecklund K, Mulkern RV, Warfield SK (2013) Reliable estimation of incoherent motion parametric maps from diffusion-weighted MRI using fusion bootstrap moves. Med Image Anal 17:325–336
While PT (2017) A comparative simulation study of Bayesian fitting approaches to intravoxel incoherent motion modeling in diffusion-weighted MRI. Magn Reson Med 78:2373–2387
Acknowledgements
The study was supported by grants from the Swedish Cancer Society and the King Gustav V Jubilee Clinic Cancer Research Foundation.
Author information
Authors and Affiliations
Contributions
Jalnefjord was responsible for study conception and design, acquisition of data, analysis, and interpretation of data, and drafting of the manuscript. Andersson, Montelius, Elf, Johanson, and Svensson were responsible for acquisition of data and critical revision of the manuscript. Starck and Ljungberg were responsible for study conception and design, analysis and interpretation of data, and critical revision of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by the regional ethical review board in Gothenburg. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individuals in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Jalnefjord, O., Andersson, M., Montelius, M. et al. Comparison of methods for estimation of the intravoxel incoherent motion (IVIM) diffusion coefficient (D) and perfusion fraction (f). Magn Reson Mater Phy 31, 715–723 (2018). https://doi.org/10.1007/s10334-018-0697-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-018-0697-5