Abstract
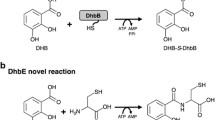
Nonribosomal peptide synthetases (NRPSs) are multi-modular enzymes involved in the biosynthesis of natural products. Bacillamide C was synthesized by Bacillus atrophaeus C89. A nonribosomal peptide synthetase (NRPS) cluster found in the genome of B. atrophaeus C89 was hypothesized to be responsible for the biosynthesis of bacillamide C using alanine and cysteine as substrates. Here, the structure analysis of adenylation domains based on homologous proteins with known crystal structures indicated locations of the substrate-binding pockets. Molecular docking suggested alanine and cysteine as the potential substrates for the two adenylation domains in the NRPS cluster. Furthermore, biochemical characterization of the purified recombinant adenylation domains proved that alanine and cysteine were the optimum substrates for the two adenylation domains. The results provided the in vitro evidence for the hypothesis that the two adenylation domains in the NRPS of B. atrophaeus C89 preferentially select alanine and cysteine, respectively, as a substrate to synthesize bacillamide C. Furthermore, this study on substrates selectivity of adenylation domains provided basis for rational design of bacillamide analogs.





Similar content being viewed by others
References
Bloudoff K, Fage CD, Marahiel MA, Schmeing TM (2017) Structural and mutational analysis of the nonribosomal peptide synthetase heterocyclization domain provides insight into catalysis. Proc Natl Acad Sci 114:95–100
Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30:1545–1614
Brooks BR, Bruccoleri RE, Olafson BD, Swaminathan S, Karplus M (1983) CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4:187–217
Challis GL, Ravel J, Townsend CA (2000) Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol 7:211–224
Conti E, Stachelhaus T, Marahiel MA, Brick P (1997) Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J 16:4174–4183
Dieckmann R, Lee YO, van Liempt H, von Dohren H, Kleinkauf H (1995) Expression of an active adenylate-forming domain of peptide synthetases corresponding to acyl-CoA-synthetases. FEBS Lett 357:212–216
Eppelmann K, Stachelhaus T, Marahiel MA (2002) Exploitation of the selectivity-conferring code of nonribosomal peptide synthetases for the rational design of novel peptide antibiotics. Biochem 41:9718–9726
Huang T, Wang Y, Yin J, Du Y, Tao M, Xu J, Chen W, Lin S, Deng Z (2011) Identification and characterization of the pyridomycin biosynthetic gene cluster of Streptomyces pyridomyceticus NRRL B-2517. J Biol Chem 286:20648–20657
Ivanova V, Kolarova M, Aleksieva K, Gräfe U, Dahse HM, Laatsch H (2007) Microbiaeratin, a new natural indole alkaloid from a Microbispora aerata strain, isolated from Livingston Island, Antarctica. Prep Biochem Biotechnol 37:161–168
Jeong S-Y, Ishida K, Ito Y, Okada S, Murakami M (2003) Bacillamide, a novel algicide from the marine bacterium, Bacillus sp. SY-1, against the harmful dinoflagellate, Cochlodinium polykrikoides. Tetrahedron Lett 44:8005–8007
Khurana P, Gokhale R, Mohanty D (2010) Genome scale prediction of substrate specificity for acyl adenylate superfamily of enzymes based on active site residue profiles. BMC Bioinform 11:1471–2105
Lee TV, Johnson LJ, Johnson RD, Koulman A, Lane GA, Lott JS, Arcus VL (2010) Structure of a eukaryotic nonribosomal peptide synthetase adenylation domain that activates a large hydroxamate amino acid in siderophore biosynthesis. J Biol Chem 285:2415–2427
Liu F, Sun W, Su F, Zhou K, Li Z (2012) Draft genome sequence of the sponge-associated strain Bacillus atrophaeus C89, a potential producer of marine drugs. J Bacteriol 194:4454
Li ZY, Hu Y, Huang YQ, Huang Y (2007) Isolation and phylogenetic analysis of the biologically active bacteria associated with three South China Sea sponges. Microbiol 76:494–499
Marshall CG, Burkart MD, Keating TA, Walsh CT (2001) Heterocycle formation in vibriobactin biosynthesis: alternative substrate utilization and identification of a condensed intermediate. Biochem 40:10655–10663
Omura S, Suzuki Y, Kitao C, Takahashi Y, Konda Y (1975) Isolation of a new sulfur-containing basic substance from a Thermo actinomyces species. J Antibiot 28:609–610
Röttig M, Medema MH, Blin K, Weber T, Rausch C, Kohlbacher O (2011) NRPSpredictor2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res 39:362–367
Sali A, Blundell T (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Schneider K, Hovel K, Witzel K, Hamberger B, Schomburg D, Kombrink E, Stuible HP (2003) The substrate specificity-determining amino acid code of 4-coumarate: CoA ligase. Proc Natl Acad Sci 100:8601–8606
Schwarzer D, Marahiel MA (2001) Multimodular biocatalysts for natural product assembly. Naturwissenschaften 88:93–101
Stachelhaus T, Mootz HD, Marahiel MA (1999) The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol 6:493–505
Socha AM, Long RA, Rowley DC (2007) Bacillamides from a hypersaline microbial mat bacterium. J Nat Prod 70:1793–1795
Tabata N, Tomoda H, Zhang H, Uchida R, Omura S (1999) Zelkovamycin, a new cyclic peptide antibiotic from Streptomyces sp. K96-0670. II. Structure elucidation. J Antibiot 52:34–39
Wu G, Robertson DH, Brooks CL, Vieth M (2003) Detailed analysis of grid-based molecular docking: a case study of CDOCKER—a CHARMm-based MD docking algorithm. J Comput Chem 24:1549–1562
Yu LL, Li ZY, Peng CS, Li ZY, Guo YW (2009) Neobacillamide A, a novel thiazole-containing alkaloid from the marine bacterium Bacillus vallismortis C89, associated with South China Sea sponge Dysidea avara. Helv Chim Acta 92:607–612
Yuwen L, Zhang F-L, Chen Q-H, Lin S-J, Zhao Y-L, Li Z-Y (2013) The role of aromatic l-amino acid decarboxylase in bacillamide C biosynthesis by Bacillus atrophaeus C89. Sci Rep 3:1753
Zhang K, Nelson KM, Bhuripanyo K, Grimes KD, Zhao B, Aldrich CC, Yin J (2012) Engineering the substrate specificity of the DhbE adenylation domain by yeast cell surface display. Chem Biol 20:92–101
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC) (31300104).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
The structures of bacillamide A, bacillamide B, bacillamide C, bacillamide D and bacillamide E
Fig. S2
NRPSpredictor2 prediction report for the A1 and A2 domains. Location of the A domain within the sequence and the bit score of the PFAM-HMM are provided. The green checkmark signals indicate that the signature sequence lies within the applicability domain. The list of predictions is provided along with the scores of the respective SVM predictors. The final row provides the nearest sequence neighbor in the NRPSpredictor2 database (based on the Stachelhaus code) and the respective sequence identity (%)
Fig. S3
Ramachandran plot atlas of the A-domain structure model (a) and Ramachandran plot atlas of the A-domain structure model after energy minimization (b). (1) A1 domain; (2) A2 domain. The figure is plotted using Discovery Studio
Rights and permissions
About this article
Cite this article
Zhang, F., Wang, Y., Jiang, Q. et al. Substrate selection of adenylation domains for nonribosomal peptide synthetase (NRPS) in bacillamide C biosynthesis by marine Bacillus atrophaeus C89. J Ind Microbiol Biotechnol 45, 335–344 (2018). https://doi.org/10.1007/s10295-018-2028-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-018-2028-2