Abstract
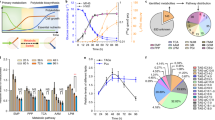
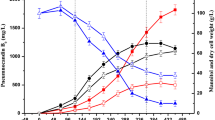
It was found that S-adenosylmethionine (SAM) could effectively improve avermectin titer with 30–60 μg/mL addition to FH medium. To clearly elucidate the mechanism of SAM on intracellular metabolites of Streptomyces avermitilis, a GC–MS-based comparative metabolomics approach was carried out. First, 230 intracellular metabolites were identified and 14 of them remarkably influenced avermectin biosynthesis were discriminative biomarkers between non-SAM groups and SAM-treated groups by principal components analysis (PCA) and partial least squares (PLS). Based on further key metabolic pathway analyses, these biomarkers, such as glucose, oxaloacetic acid, fatty acids (in soybean oil), threonine, valine, and leucine, were identified as potentially beneficial precursors and added in medium. Compared with single-precursor feeding, the combined feeding of the precursors and SAM markedly increased the avermectin titer. The co-feeding approach not only directly verified our hypothesis on the mechanism of SAM by comparative metabolomics, but also provided a novel strategy to increase avermectin production.





Similar content being viewed by others
References
Beyoğlu D, Idle JR (2013) Metabolomics and its potential in drug development. Biochem Pharmacol 85(1):12–20
Bo T, Liu M, Zhong C, Zhang Q, Su QZ, Tan ZL, Han PP, Jia SR (2014) Metabolomic analysis of antimicrobial mechanisms of ε-poly-l-lysine on Saccharomyces cerevisiae. J Agric Food Chem 62(19):4454–4465
Bolten CJ, Wittmann C (2008) Appropriate sampling for intracellular amino acid analysis in five phylogenetically different yeasts. Biotechnol Lett 30(11):1993–2000
Chen TS, Inamine ES (1989) Studies on the biosynthesis of avermectins. Arch Biochem Biophys 270(2):521–525
Dietmair S, Hodson MP, Quek LE, Timmins NE, Chrysanthopoulos P, Jacob SS, Gray P, Nielsen LK (2012) Metabolite profiling of CHO cells with different growth characteristics. Biotechnol Bioeng 109(9):1404–1414
Dikbas N (2010) Determination of antibiotic susceptibility and fatty acid methyl ester profiles of Bacillus cereus strains isolated from different food sources in Turkey. Afr J Biotechnol 9(11):1641–1647
Ding MZ, Zhou X, Yuan YJ (2010) Metabolome profiling reveals adaptive evolution of Saccharomyces cerevisiae during repeated vacuum fermentations. Metabolomics 6(1):42–55
Dotzlaf JE, Metzger LS, Foglesong MA (1984) Incorporation of amino acid-derived carbon into tylactone by Streptomyces fradiae GS14. Antimicrob Agents Chemother 25(2):216–220
Fang ZZ, Tosh DK, Tanaka N, Wang H, Krausz KW, O’Connor R, Jacobson KA, Gonzalez FJ (2015) Metabolic mapping of A3 adenosine receptor agonist MRS5980. Biochem Pharmacol 97(2):215–223
Gao H, Liu M, Liu J, Dai H, Zhou X, Liu X, Zhuo Y, Zhang W, Zhang L (2009) Medium optimization for the production of avermectin B1a by Streptomyces avermitilis 14-12A using response surface methodology. Bioresour Technol 100(17):4012–4016
Gao H, Liu M, Zhou X, Liu J, Zhuo Y, Gou Z, Xu B, Zhang W, Liu X, Luo A, Zheng C, Chen X, Zhang L (2010) Identification of avermectin-high-producing strains by high-throughput screening methods. Appl Microbiol Biotechnol 85(5):1219–1225
Guo G, Tian PP, Tang D, Wang XX, Yang HJ, Cao P, Gao Q (2015) Metabolomics analysis between wild-type and industrial strains of Streptomyces avermitilis based on gas chromatography-mass spectrometry strategy. Lect Notes Electr Eng 333:477–485
Hesse H, Kreft O, Maimann S, Zeh M, Willmitzer L, Höfgen R (2001) Approaches towards understanding methionine biosynthesis in higher plants. Amino Acids 20(3):281–289
Huh JH, Kim DJ, Zhao XQ, Li M, Jo YY, Yoon TM, Shin SK, Yong JH, Ryu YW, Yang YY, Suh JW (2004) Widespread activation of antibiotic biosynthesis by S-adenosylmethionine in streptomycetes. FEMS Microbiol Lett 238(2):439–447
Ikeda H, Kotaki H, Tanaka H, Ōmura S (1988) Involvement of glucose catabolism in avermectin production by Streptomyces avermitilis. Antimicrob Agents Chemother 32:282–284
Ikeda H, Ōmura S (1997) Avermectin biosynthesis. Chem Rev 97(7):2591–2610
Khaoua S, Lebrihi A, Laakel M, Schneider F, Germain P, Lefebvre G (1992) Influence of short-chain fatty acids on the production of spiramycin by Streptomyces ambofaciens. Appl Microbiol Biotechnol 36(6):763–767
Korneli C, Bolten CJ, Godard T, Franco-Lara E, Wittmann C (2012) Debottlenecking recombinant protein production in Bacillusmegaterium under large-scale conditions-targeted precursor feeding designed from metabolomics. Biotechnol Bioeng 109(6):1538–1550
Lee JJ, Chen L, Shi J, Trzcinski A, Chen WN (2014) Metabolomic profiling of Rhodosporidium toruloides grown on glycerol for carotenoid production during different growth phases. J Agric Food Chem 62(41):10203–10209
Li L, Guo J, Wen Y, Chen Z, Song Y, Li J (2010) Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains. J Ind Microbiol Biotechnol 37(7):673–679
Liu X, Lu YF, Guan X, Zhao M, Wang J, Li F (2016) Characterizing novel metabolic pathways of melatonin receptor agonist agomelatine using metabolomic approaches. Biochem Pharmacol 109:70–82
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Mrozik HH (1983) Processes for the interconversion of avermectin compounds. US Patent 4,423,209
Novák J, Hájek P, Řezanka T, Vaněk Z (1992) Nitrogen regulation of fatty acids and avermectins biosynthesis in Streptomyces avermitilis. FEMS Microbiol Lett 72(1):57–61
Ochi K, Saito Y, Umehara K, Ueda I, Kohsaka M (1984) Restoration of aerial mycelium and antibiotic production in a Streptomyces griseoflavus arginine auxotroph. J Gen Microbiol 130(8):2007–2013
Okamoto S, Lezhava A, Hosaka T, Okamoto-Hosoya O, Ochi K (2003) Enhanced expression of S-adenosylmethionine synthetase causes overproduction of actinorhodin in Streptomyces coelicolor A3(2). J Bacteriol 185(2):601–609
Ōmura S, Ikeda H, Tanaka H (1991) Selective production of specific components of avermectins in Streptomyces avermitilis. J Antibiot (Tokyo) 44(5):560–563
Schulman MD, Acton SL, Valentino DL, Arison BH (1990) Purification and identification of dTDP-oleandrose, the precursor of the oleandrose units of the avermectins. J Biol Chem 265(8):16965–16970
Shin SK, Xu D, Kwon HJ, Suh JW (2006) S-adenosylmethionine activates adpA transcription and promotes streptomycin biosynthesis in Streptomyces griseus. FEMS Microbiol Lett 259(1):53–59
Sima C, Dougherty ER (2006) What should be expected from feature selection in small-sample settings. Bioinformatics 22(19):2430–2436
Szeto SS, Reinke SN, Sykes BD, Lemire BD (2010) Mutations in the Saccharomyces cerevisiae succinate dehydrogenase result in distinct metabolic phenotypes revealed through 1H NMR-based metabolic footprinting. J Proteom Res 9(12):6729–6739
Tang L, Zhang YX, Hutchinson CR (1994) Amino acid catabolism and antibiotic synthesis: valine is a source of precursors for macrolide biosynthesis in Streptomyces ambofaciens and Streptomyces fradiae. J Bacteriol 176(19):6107–6119
Tian X, Wang Y, Chu J, Zhuang Y, Zhang S (2016) Enhanced L-lactic acid production in Lactobacillus paracasei by exogenous proline addition based on comparative metabolite profiling analysis. Appl Microbiol Biotechnol 100(5):2301–2310
Van Voorhisa WC, Hooft van Huijsduijnend R, Wells TN (2015) Profile of William C. Campbell, Satoshi Ōmura, and Youyou Tu, Nobel laureates in physiology or medicine. Proc Natl Acad Sci USA 112(52):15773–15776
Wang B, Jiu J, Liu H, Huang D, Wen J (2015) Comparative metabolic profiling reveals the key role of amino acids metabolism in the rapamycin overproduction by Streptomyces hygroscopicus. J Ind Microbiol Biotechnol 42(6):949–963
Wold S, Geladi P, Esbensen K, Öhman J (1987) Multi-way principal components-and PLS-analysis. J Chemometr 1(1):41–56
Wu C, Choi YH, van Wezel GP (2016) Metabolic profiling as a tool for prioritizing antimicrobial compounds. J Ind Microbiol Biotechnol 43(2–3):299–312
Xu Z, Cen P (1999) Stimulation of avermectin B1a biosynthesis in Streptomyces avermilitis by feeding glucose and propionate. Biotechnol Lett 21(1):91–95
Yin P, Li YY, Zhou J, Wang YH, Zhang SL, Ye BC, Ge WF, Xia YL (2013) Direct proteomic mapping of Streptomyces avermitilis wild and industrial strain and insights into avermectin production. J Proteom 79:1–12
Yoon GS, Ko KH, Kang HW, Suh JW, Kim YS, Ryu YW (2006) Characterization of S-adenosylmethionine synthetase from Streptomyces avermitilis NRRL8165 and its effect on antibiotic production. Enzyme Microb Technol 39(3):466–473
Yoon YJ, Kim ES, Hwang YS, Choi CY (2004) Avermectin: biochemical and molecular basis of its biosynthesis and regulation. Appl Microbiol Biotechnol 63(6):626–634
Acknowledgements
This work was financially supported by the National Basic Research Program (973 Program) of China (2013CB734004), and the Natural Science Foundation of China (31370075, 31471725, 61603273), and the Youth Innovation Fund of Tianjin University of Science and Technology of China (2014CXLG28). We also appreciate Dr. Arnold L. Demain for his valuable advice with the manuscript, Dr. Lixin Zhang for providing the experimental strain and Mr. Gang Guo for his technical support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10295_2016_1883_MOESM1_ESM.ppt
Fig. 1S PCA score plots of different SAM additions at different timepoints. PCA score plots (PC1, 72.35% of total variance; PC2, 21.47% of total variance). In the score plots, the confidence interval is defined by Hotelling’s T2 ellipse (95% confidence interval), and observations outside the ellipse are considered outliers (PPT 160 kb)
10295_2016_1883_MOESM2_ESM.pptx
Fig. 2S PLS score plots (a, c, e) and loading plots (b, d, f) of samples with different contents of SAM. Sampling time (a, b) at 24 h, (c, d) at 48 h, and (e, f) at 72 h. In the score plots, the confidence interval is defined by Hotelling’s T2 ellipse (95% confidence interval), and observations outside the ellipse are considered outliers. Green icons represent the non-SAM groups; blue icons represent the 30 μg/mL SAM-treated groups; and red icons represent the 60 μg/mL SAM-treated groups (PPTX 134 kb)
10295_2016_1883_MOESM3_ESM.pptx
Fig. 3S PLS score plots (a, c, e) and loading plots (b, d, f) of samples with different contents of SAM. Sampling time (a, b) at 24 h, (c, d) at 48 h, and (e, f) at 72 h. In the score plots, the confidence interval is defined by Hotelling’s T2 ellipse (95% confidence interval), and observations outside the ellipse are considered outliers. Green icons represent the non-SAM groups; blue icons represent the 30 μg/mL SAM-treated groups; and red icons represent the 60 μg/mL SAM-treated groups (PPTX 464 kb)
Rights and permissions
About this article
Cite this article
Tian, P., Cao, P., Hu, D. et al. Comparative metabolomics reveals the mechanism of avermectin production enhancement by S-adenosylmethionine. J Ind Microbiol Biotechnol 44, 595–604 (2017). https://doi.org/10.1007/s10295-016-1883-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1883-y