Abstract
Recent emerging hybrid technology of positron emission tomography/magnetic resonance (PET/MR) imaging has generated a great need for an accurate MR image-based PET attenuation correction. MR image segmentation, as a robust and simple method for PET attenuation correction, has been clinically adopted in commercial PET/MR scanners. The general approach in this method is to segment the MR image into different tissue types, each assigned an attenuation constant as in an X-ray CT image. Machine learning techniques such as clustering, classification and deep networks are extensively used for brain MR image segmentation. However, only limited work has been reported on using deep learning in brain PET attenuation correction. In addition, there is a lack of clinical evaluation of machine learning methods in this application. The aim of this review is to study the use of machine learning methods for MR image segmentation and its application in attenuation correction for PET brain imaging. Furthermore, challenges and future opportunities in MR image-based PET attenuation correction are discussed.
Similar content being viewed by others
Introduction
Positron emission tomography (PET) is an imaging modality that provides direct imaging of physiological biomarkers using radiolabeled gamma-ray emitting molecules. The knowledge of the tissue-dependent attenuation map, needed for attenuation correction, is a critical step to achieve an accurate PET image reconstruction. Figure 1 shows the effect of attenuation correction on reconstructed PET images. The attenuation map is usually obtained by performing an additional scan using X-ray computed tomography (CT) [1]. CT image intensity measured in the Hounsfield unit is a map of the normalized X-ray attenuation coefficients, which reflects the anatomical, physiological and pathological states of the underlying tissues. Therefore, the CT image can be mathematically converted to the equivalent linear X-ray attenuation coefficients [2, 3]. Since X-rays and gamma-rays have similar attenuations in biological tissues, X-ray CT is the most straightforward way for PET attenuation correction. However, it introduces additional ionizing radiations to the imaging subjects.
A reconstructed PET image without attenuation correction (a), and with attenuation correction (b) using the [(18)F]-fluorodeoxyglucose ((18)F-FDG) radiotracer. Adopted form [4]
On the other hand, magnetic resonance (MR) imaging is nowadays considered the premier modality for imaging the brain structures and functions due to its excellent soft tissue contrast, high spatial resolution, and lack of ionizing radiation. Therefore, MR images have been extensively used for visualizing, analyzing, diagnosis, treatment planning, and follow-up of a variety of neurological conditions.
To take advantages of both MRI and PET, hybrid PET/MR systems were recently introduced and applied in the clinical molecular imaging applications [5]. However signal intensity in MR images is not directly correlated to attenuation coefficient which is required for attenuation correction in PET image reconstructions [2]. Therefore, MR image-based attenuation correction has become one of the challenges in PET/MR systems [6]. There are different approaches are used to addressing this challenge which are discussed in “MR Image-Based Attenuation Correction for Brain PET Imaging”. T1-weighted and T2-weighted MR images are commonly used in MR image-based attenuation correction. Other MR image pulse sequences which provide more details in morphological information can also be used for this purpose. These include diffusion-weighted images (DWI) [7], short echo time (STE) [8], ultra-short echo time (UTE) [9], zero echo time (ZTE) [10], dynamic contrast-enhanced (DCE) imaging [11], and magnetization-prepared rapid acquisition gradient echo (MP-RAGE) sequences [12].
Current research utilizes different machine learning techniques and MR image data acquisition sequences to perform MR image segmentation for different medical applications, including PET/MR attenuation correction. DWI sequence [13] or a combination of sequences [14, 15] is the most routinely used for the diagnosis and follow-up in ischemic and hemorrhagic stroke. For quantitative analysis in multiple sclerosis, T2-weighted MR images is commonly used sequence either as a single imaging sequence [16] or in a multi-sequence approach [17,18,19]. T1-weighted MR images are frequently used to assess biomarkers of Alzheimer’s disease such as hippocampal atrophy, ventricle enlargement and cortex shrinkage [20, 21]. Brain tumor segmentation of MR images received much attention over the last decade, especially for treatment planning and follow-up. A range of MR image sequences were used as input to segmentation procedure: single MR image sequence with [22] and without [23] contrast agent, or multi-sequence MR images with [24,25,26,27] or without contrast [24, 28].
The process of segmentation is performed by segmenting the brain MR images into three main tissue classes: white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) as well as the lesion regions. These have been a large body of literatures on brain image segmentation methods as thresholding, edge based, watershed based, and various machine learning techniques. For instance, random forest classifier has been used for diagnosis of Alzheimer disease by biomarkers such as segmenting white matter lesions [29] or hippocampus for diagnosis [30], and for segmentation of brain tumor lesions [31]. Support vector machine (SVM) showed good segmentation results in white matter lesion [32], multiple sclerosis region [33], brain tumor lesions [24] or to diagnose Alzheimer disease [20]. Additionally, probabilistic models were proposed to efficiently segment the brain tissue into three classes or to only extract the region of interest by applying the inverted Dirichlet mixture Model [34], Markov random field [35] and Bayesian model [36]. Furthermore, neural network such as multilayer perceptron [37], self-organizing map (SOM) network [38], extreme learning machine (ELM) [39], and cellular neural network [40] achieved also good results in segmenting the brain into different regions. Unsupervised learning, specifically clustering techniques, have been widely used for skull striping [41], white matter lesion segmentation [42], and tumor region segmentation [23]. Recently, deep learning proved powerful performance in several brain segmentation applications using convolutional neural networks (CNN) for skull stripping [43], stroke lesion segmentation [44], multiple sclerosis segmentation [45], and brain tumor segmentation [27].
The aim of this literature review is to highlight the recent progress made on MR image-based PET attenuation correction using machine learning for segmentation of brain tissues. The structure of this review is as follows: Section “MR Image-Based Attenuation Correction for Brain PET Imaging” introduces the MR image-based attenuation correction for brain PET imaging. Section “Machine Learning-Based Segmentation Methods for PET Attenuation Correction in Brain Imaging” represents the machine learning-based segmentation methods for PET attenuation correction in brain imaging. Section “Performance Metrics and Clinical Evaluation” discusses the evaluation metrics and clinical evaluation. The challenges and opportunities are discussed in “Challenges and Future Opportunities” and the conclusion is presented in “Conclusion”.
MR Image-Based Attenuation Correction for Brain PET Imaging
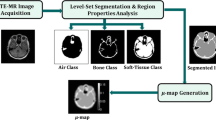
With the recent introduction of hybrid PET/MR scanners, PET attenuation correction maps need to be generated from MR images. Unlike CT, the MR image signal intensity has no direct mapping to PET attenuation coefficients. For instance, bone and air have similar intensity values in conventional MR images while they have quite different attenuation coefficients [3]. Figure 2 illustrates the difference between CT imaging-based and MR imaging-based attenuation correction.
Generating pseudo CT from MR images requires segmented MR images where tissue classes assigned a specific attenuation coefficient. Moreover, clinically adopted method for MR image-based attenuation correction, also available for commercial PET/MR scanners, involves MR images segmentation as a basis to generate pseudo CT images [46]. MR images are commonly segmented into three or more tissue classes then predefined attenuation coefficients will be assigned to each voxel tissue class. Hence, there is a great need for a reliable method to generate the attenuation correction coefficients from MR images. As shown, it requires complex transformation to produce pseudo CT from MR images. The main complex transformations to map a MR image to a pseudo CT images are as follows: segmentation, atlas, and machine learning which are discussed in the following paragraphs.
The segmentation methods are traditionally considered the most robust and simple methods adopted in clinical domain for MR image-based attenuation correction for PET images [47,48,49]. The first clinical hybrid PET/MR system uses the two-point Dixon gradient echo sequence which simplifies the segmentation of MR images into different tissue classes [4]. Nowadays, the commercial PET/MR systems segment images into three or four tissue classes [50], with the voxels of each tissue class assigned an approximated predefined linear attenuation coefficient [51] producing the attenuation correction map. MR image segmentation was performed using different approaches starting with simple techniques such as level set [52, 53], thresholding [50, 54,55,56,57,58,59,60], and radon transform [59] until more complicated techniques such as clustering [61,62,63], classification [64] and deep learning [65,66,67]. Table 1 illustrates different segmentation methods applied to different MR image sequences. The main challenge of segmentation is the accurate delineation of bone tissue [47]. Furthermore, there is also a disagreement on the value of a linear attenuation coefficient to be assigned for the bone tissue [54].
The atlas-based methods, also referred to as the registration-based methods, involve image registration between atlas/template images (MR/CT image pairs) and the target MR image using nonlinear transformation. First, the MR image of the subject is co-registered with the atlas MR image. Then, the obtained transformation is applied to atlas CT image to create subject specific attenuation correction map [48]. The quality of PET reconstruction is highly dependent on the registration algorithms accuracy. Different atlas-based techniques were proposed in the literature [46, 68,69,70]. Most of the atlas-based methods use machine learning to estimate the pseudo CT image using MR image features such as signal intensity and geometric metrics to learn the relationship between MR signal and Hounsfield units in CT. This method is time consuming and potentially have decreased accuracy under anatomical variations, especially in cases undergoing neurosurgery [71].
Machine learning methods are related to both segmentation- and atlas-based methods where different algorithms are applied either to perform MR image segmentation [61,62,63,64, 72, 73] or post-registration process to learn the complex mapping from MR images to CT in order to generate the pseudo CT images [74,75,76,77,78,79,80,81,82,83]. Different machine learning techniques have been applied such as Gaussian mixture regression model, k-nearest neighbors (kNN) regression, random forest classifier, neural networks, clustering techniques, and deep learning. In the next section, MR images segmentation methods using machine learning for brain PET attenuation correction are reviewed in detail. These include image clustering, image classification, and deep learning.
Machine Learning-Based Segmentation Methods for PET Attenuation Correction in Brain Imaging
Tables 2 and 3 summarize the three main categories of machine learning techniques proposed for MR images segmentation: clustering, classification, and deep learning.
Clustering
Khateri et al. [62] used a combination of STE sequences with 2-point Dixon technique along with a fuzzy C-means (FCM) clustering-based segmentation method to detect bone tissue. They segmented the brain into four clusters, namely cortical bone, soft tissue, adipose tissue, and air. They concluded that the clustering technique is an appropriate approach to segment bone and air in the sinusoidal area. The bone segmentation results achieved more than 90% in terms of accuracy, sensitivity, and specificity. However, the eye area can be misclassified as bone. The results were validated using manually segmented bone regions on STE MR images by a neuroradiologist expert. Later, the same team (Khateri et al. [8]) applied FCM clustering to segment the brain into three tissue classes (cortical bone, soft tissue, and air) using the same combination of MR sequences (STE + Dixon). They used morphologic operations as post segmentation to reduce susceptibility error. This method was evaluated with CT-based attenuation correction maps as shown in Fig. 3. The visual comparison showed the high similarity between MR and CT segmentation results. The evaluation measures are signal-to-noise ratio (SNR), accuracy, sensitivity, specificity, and correlation plots. The segmentation results proved that the combination of STE sequence with a clustering technique is a potential alternative for UTE sequences for PET attenuation correction. They concluded from the obtained attenuation correction maps that the ethmoid sinuses are the most error-prone areas with the largest difference in the paranasal area.
MR image segmentation results achieved by [8] using clustering technique with a the reference CT images, b the segmented MR images, and c the difference between the two modalities
Fei et al. [63] developed a multiscale segmentation approach using radon transform of T1-weighted MR images to segment the head image into skull, scalp, and brain tissue. Then, the brain tissue was classified into three classes: GM, WM, and CS by applying unsupervised clustering technique. The images were firstly processed with anisotropic diffusion filter to construct a multiscale image series in order to overcome the blurred edges drawback. Then, a multiscale FCM technique was applied to allow multiscale processing from the coarse to fine levels. Afterwards, predefined attenuation coefficients were assigned to each tissue class. The segmentation is evaluated using dice similarity measurement. The quality of PET images is compared with transmission (TX)-based attenuation correction by visual inspection followed by quantitative measurements which are relative difference, mean squared error (MSE), and peak signal-to-noise ratio (PSNR). The overlap ratio between the segmented and ground truth CT is around 85% and the difference between the MR- and TX-based attenuation correction maps is less than 7%.
Su et al. [72] proposed an attenuation correction method using a single acquisition, undersampled UTE-mDixon, MR images. They applied FCM clustering algorithm to segment the head into five different tissue classes including brain, air, fat, fluid, and bone. After optimizing MR images, three image features which are Dixon-fat, Dixon-water, and R2 were used as input to the unsupervised clustering algorithm. The segmented MR voxels were assigned attenuation correction coefficients to generate the pseudo CT images. The coordinates of the centroids of different tissue types were calculated to evaluate the segmentation results. Then, the obtained pseudo CT images are compared with measured low-dose CT images visually and subjectively by calculating the CT histogram and mean absolute predication deviation (MAPD).
Classification
Shi et al. [4] proposed bone refinement method for existing attenuation correction map. This method started with an existing attenuation correction map from the vendor or obtained from any segmentation-based method then refined the attenuation correction map gradually by learning the relationship between the MR image and the attenuation correction map. The learning process performed using multiresolution regional learning approach by applying SVM classifier to refine the attenuation correction coefficients using training features of UTE1, UTE2, and MP-RAGE sequences. The resulting attenuation correction map was compared with vendor map and CT-based attenuation correction map by measuring the bone recovery rate, dice coefficient, voxel-wise error, and region-wise error. The measurements showed that the proposed method enhanced the attenuation coefficient map. The reconstructed PET images displayed that the error is larger in the regions near the skull. They also concluded from the visual inspection that the proposed method reduced the underestimation of PET activities. However, the results are still not comparable with atlas-based methods.
Santos et al. [61] performed a learning-based segmentation method of the skull using probabilistic feed-forward neural network which requires user interaction. UTE sequences were used to segment the brain into four classes: air, brain + soft tissue, CSF, and bone. The model was trained using two patches of raw MR intensities (UTE1 and UTE2) as input features. This method was compared with CT-based attenuation correction map by calculating the accuracy, the dice similarity score, and the visual comparison. They found that this approach achieved high values for the co-classification of air and soft tissue. However, the bone values are low. This algorithm depends on the patient’s MR intensity values which affects the performance if there is a big difference between patients’ MR intensities.
Chan et al. [64] proposed a segmentation method based on tissue classification to differentiate bone from air. Brieman’s random forest classifier was trained using a set of features include gradient, textural, and contextual features extracted from MP-RAGE MR sequences and uncorrected PET images. The segmentation results of MP-RAGE images are shown in Fig. 4. Dice similarity score for each tissue class, accuracy, area under the curve (AUC) of the receiver operating characteristic (ROC) curve, and visual assessment were used to evaluate the segmentation performance. The evaluation metrics were compared with CT-based classification as ground truth. They concluded that the segmentation results were improved when including the uncorrected PET features compared with segmentation using MR image features only.
MR image segmentation result achieved by [64] using voxel classification to differentiate bone from air. a T1-weighted MR image, b segmented MR image, and c corresponding CT image as ground truth
Koesters et al. [73] applied Adaboost classifier to extract bone tissue from T1-weighted MR sequences. Afterwards, the bone attenuation coefficient was added to Dixon-based attenuation map from the manufacturer that reflects four tissue attenuation coefficients but not bone tissue. This method was evaluated by comparing the standardized uptake value (SUV) estimation between CT, Dixon, and the proposed model for whole-brain and regional analyses. The results showed that there is a significant improvement in terms of SUV estimation bias by comparing the proposed model with Dixon-based MR images. The proposed approach reduced the whole brain SUV estimation bias of Dixon-based approach by 95% and a similar residual SUV bias to CT-based approach.
Deep Learning
Deep learning is an emerging technology in machine learning which represents advanced and more complex forms of neural networks. Deep networks are self-learning structures capable of learning high-level image features and modeling a nonlinear mapping between different image spaces through the convolution process. These methodologies showed their superiority in several medical applications, which paved the way for their recent exploration for PET attenuation correction by generating attenuation maps using different MR sequences.
There are only few publications that applied deep learning to brain MR image segmentation for PET attenuation corrections [65,66,67]. Each of these proposed studies used different MR image sequences and network architecture to train the deep network.
Liu et al. [65] applied a deep convolutional encoder-decoder network called Segnet [85] using T1-weighted MR images. This work required a co-registration between CT and MR images before the training process and the creation of ground truth. To train the network, the labels were generated by segmenting the CT images into three classes (air, soft tissue, and bone) using intensity-based thresholding technique. These classes were assigned an attenuation coefficient value to generate the pseudo CT images as illustrated in Fig. 5. Dice similarity score for each class was calculated to evaluate the segmentation results and PET reconstruction error was measured to quantify the obtained PET image. The proposed method was compared with Dixon-based soft tissue and air segmentation and anatomic CT-based template registration. The results achieved accurate pseudo CT scans and good PET images with lower errors compared with Dixon-based and CT-based attenuation correction. The main limitation of this approach is the intrasubject registration which would affect the segmentation performance.
a Pseudo CT image obtained by segmenting b T1-weighted MR image with the use of c CT image as a ground truth [65]
Jang et al. [66] used UTE and out of phase (fat and water) MR images which were acquired using dual echo ramped hybrid encoding (dRHE) to segment the brain into three classes: air, soft tissue, and bone. UTE sequences used as an input to retrain a pretrained deep network [86] with T1-weighted MR images. Transfer learning was applied to adopt the knowledge learnt from other MR sequence to UTE sequences to improve the learning and obtain a reliable training. The obtained segmented MR images were processed using conditional random field technique to refine the segmentation results. Furthermore, the out of phase images were used to segment the soft tissue into fat and water components using two-point Dixon-based segmentation. The segmented labels from deep learning- and Dixon-based water and fat images were integrated to generate the pseudo CT images. The proposed method was compared with three other MR-based attenuation correction methods while using CT-based attenuation correction map as the standard reference. Dice similarity score between the predicted labels and CT ground truth images was calculated to evaluate the segmentation results. The corrected PET images were evaluated using relative PET errors. The results showed that this method is clinically feasible with rapid dRHE acquisition time and less than 1% relative PET error in most brain regions. They also showed that the application of conditional random field did not increase the computation time but improved the air and bone detection as well as it corrected some artifacts.
Arabi et al. [84] proposed a deep learning generative adversarial semantic model that generates pseudo CT images for MR image-based attenuation correction. The generative adversarial network consists of two main components: synthesis network and segmentation network. The synthesis part generates pseudo CT images from T1-weighted MR images and the segmentation network segments the obtained pseudo CT images into four tissue classes which are bone, air, soft tissue, and background. The two blocks are connected to each other since the segmentation network contributes to the backpropagation process on the synthesis network. The method was compared with an atlas-based method and a commercial segmentation based method by calculating the cortical bone dice similarity coefficient, mean error, mean absolute error, SUV error, relative mean square error (RMSE), PSNR, and structural similarity index measurement (SSIM). They found the proposed method and atlas-based method have similar performance with tolerable errors. They also concluded that the deep learning method outperforms the commercial segmentation-based approach used in the clinic. CT-based attenuation correction maps were used as standard reference for evaluation.
Performance Metrics and Clinical Evaluation
Performance Metrics
The segmentation accuracy is crucial for medical image analysis and quantification. Usually, the segmentation is evaluated using different evaluation metrics such as dice similarity coefficient or F1- measure [63], Jaccard index [87], sensitivity (recall) [62], specificity (precision) [62], accuracy [64], AUC-ROC [64], and false discovery rate [4].
The attenuation correction map is usually evaluated by calculating the PET reconstruction error [56], MAPE [58], RMSE [57], relative difference maps [63], and visual comparison of the maps [61]. The majority of studies used CT attenuation correction map as the gold standard reference for validation [52,53,54,55,56,57,58].
Clinical Evaluation of PET Attenuation Correction Maps
A variety of clinical studies were carried out to evaluate the clinical performance of MR image-based PET attenuation correction methods. Tables 4 and 5 summarize the clinical studies with their evaluation metrics.
For example, Aasheim et al. [88] evaluated the performance of the most recent version of Siemens UTE-based attenuation correction for PET data using seven lymphoma and twelve lung cancer patients. They concluded that further improvement is needed for accurate segmentation of bone. Choi et al. [89] studied the clinical quantification of PET using UTE-based attenuation correction including bone segmentation. They found that UTE-based attenuation correction causes spatial bias in PET quantification.
Delso et al. [90] were the first to publish a clinical evaluation of bone identification based on brain ZTE sequences. They reviewed the attenuation maps from 15 clinical datasets acquired with a PET/CT/MR trimodality setup and they found out that ZTE images are an efficient imaging sequence to overcome the limitation of bone tissue in attenuation correction maps with sufficient accuracy. Moreover, the evaluation results showed that ZTE sequence is better than UTE according to Jaccard distance value. Sekine et al. [91] proposed a study to evaluate the clinical feasibility of ZTE-based attenuation correction compared with a clinical applied method based on atlas attenuation correction. The calculations showed that the absolute relative difference between PET images is improved with ZTE-based attenuation correction. They concluded that this method is more accurate than clinical atlas attenuation correction.
Anazodo et al. [92] evaluated the addition of bone information on Dixon attenuation correction maps from T1-weighted MR images and the results proved the improvement of underestimation of PET activity. Andersen et al. [93] assessed the regional and absolute bias introduced from neglecting bone using different Dixon image-based methods. They concluded that further improvement for the existing methods is required to adopt PET/MR imaging in clinical routine.
Dickson et al. [94] assessed the quantitative accuracy of Dixon- and UTE-based MR image attenuation correction methods which were compared with CT-based attenuation correction method. Significant underestimations of activity concentrations were found using both Dixon and UTE sequences. The underestimation using UTE is less than with Dixon attenuation correction.
Baran et al. [95] developed a segmentation-based method which compared with three other MR-based methods. The proposed method is based on gaussian mixture segmentation with two different approaches of attenuation coefficients assignments (constant and continuous values). This study firstly compared two attenuation coefficients reference maps using manually segmented MR image- and CT-based maps and concluded that there is a very small mean differences across all subjects. Then, the proposed method was evaluated with CT-based attenuation coefficients and compared with the vendor UTE and UCL methods [68]. They found out that the reconstructed PET with continuous attenuation coefficients has a better agreement with the reference map especially in the cortical bones region while UCL method showed an overestimation for all brain regions. They also observed that the significant differences appear in the cerebellum region. Moreover, the segmentation results showed an underestimation in the esophagus region.
Finally, Ladefoged et al. [96] published a comparison study that evaluates eleven selected MR attenuation correction methods from the literature to study their feasibility in the clinical domain. The evaluated methods are two vendor-implemented using Dixon and UTE sequences, five atlas-based methods, one emission-based method, and three segmentation-based methods. They used a large dataset which contains 359 patients. The main finding of this study is all methods do not exceed more than 5% relative error in the whole brain and in all brain regions compared with CT-based reference map. They also found out that three template-based methods [68, 70, 97] and one segmentation-based method [50] outperform the others in terms of robustness, outliers, and clinical feasibility. Another conclusion is the vendor-implemented methods outperform other methods in term of processing time. The main limitation of this study is the results cannot be generalized since the subject datasets do not include children and patients with anatomical changes.
Challenges and Future Opportunities
MR Image Segmentation: Challenges and Opportunities
Although MR images can provide needed information for PET attenuation correction by the aforementioned methods such as tissue segmentation, MR image segmentation itself is challenging. A common problem with segmentation techniques is the misclassification of the pixels which makes the determination of boundaries very challenging. This may lead to false negatives where the lesion regions classified as healthy and false positive where the healthy regions identified as diseased lesions. Hence, robust and accurate segmentation techniques are required to be used in clinical routine.
Machine learning techniques such as neural networks, clustering, random forest, and SVM presented good accuracy with different sets of features. The combination of different techniques for different medical applications leads to better performance and accuracy such as combining random forest classifier with Markov random field to segment white matter lesions in contrast-enhanced FLAIR MR images [29] and combining regularized SVM with a kNN classifier for hippocampus segmentation as represented in [98].
Comparison between the performance and efficiency of each technique is generally applied within the same context and same medical application. For instance, there are several studies that compared the segmentation results using different classifiers as represented in [19, 99,100,101,102,103]. The comparison study between different types of classifiers showed the superiority of deep neural networks and especially CNN in the segmentation task.
The conventional machine learning workflow composes of features extraction and selection then classification. However, the feature vectors control the performance of segmentation methods rather than the classifier’s type. Therefore, more attention on the development of the feature extraction techniques should be carried out with the usage of simple classifiers. Deep learning is the best solution to avoid the hand-crafted features as the training process of the deep network learns the features automatically through the convolutional layers. At the end of the learning process, only high-level features that represent the main characteristics of the data are preserved.
Although deep learning has shown to be superior than any other machine learning techniques, it needs a good estimation of the numerous parameters of the network as well as it needs excessive training time to obtain the weights which will be used for predictions.
MR Image-Based Attenuation Correction: Challenges and Opportunities
Hybrid PET/MR scanners introduced the complementary nature of MR and PET images to clinical applications and have improved the PET quantification for disease diagnosis and treatment planning by providing different tissue characteristics. Furthermore, it also reduced the acquisition time in case of simultaneous acquisitions. The mergence of MR image modality into the field of PET attenuation correction and quantification raised the new challenges and difficulties.
Table 2 summarizes two types of segmentation methods which are applied on PET attenuation correction: supervised and unsupervised techniques. Fuzzy c-means clustering is the only unsupervised technique which is applied in the literature. However, most of these studies [8, 62] are evaluating the segmentation results by reporting the accuracy values which leads to an inaccurate evaluation since the accuracy metric is not applicable in the case of class unbalancing issue. The high values of accuracy do not indicate a robust segmentation result. On the other hand, the supervised machine learning techniques are as follows: SVM, random forest, and adaptive boosting classifiers. The random forest-based approach [64] used PET features combined with other MR image features and achieved the dice value 0.98. This method is not comparable with other methods that rely on MR features only. Moreover, the reviewed methods used different MR sequences; hence, there is no way to compare the results. Some studies used simple and conventional MR sequences such as T1-w [63, 65, 73] and others used more sophisticated sequences such as UTE sequences [61, 66].
The main limitation of deep learning-based method is the availability of big data especially with medical datasets. The deep models require a lot of data to be well trained and tested. Sufficient training data with different abnormalities should build deep learning models that can perform better than atlas-based methods which assume healthy tissue of each patient. Furthermore, the computation time of large datasets is another challenge of training deep models. The training process can take several days; hence, the usage of high performance computers and graphical processing units is mandatory. The promising thing is once the model is trained, the predication time is very short. Another common limitation in the studies that applied deep learning for PET attenuation correction is the need to apply co-registration between MR and CT images. The mis-registration can lead to further errors in the segmentation and PET reconstructions processes. Moreover, the loss function that calculates the training error can cause issues especially in the case of data unbalancing where some classes are minorities such as the bone class in the brain. Therefore, the prediction will be biased toward the majority classes and cause high specificity and low sensitivity segmentation. Voxel-wise methods [104], class weighting [18] and customized loss functions [105, 106] are some potential solutions.
MR image-based segmentation methods are the clinically adopted method in commercial scanners for attenuation correction [47]. These methods are easy to implement with low computational cost. However, they suffer from a poor segmentation by misclassifying bone (by air or soft tissue) due to a low T2 relaxation time. Consequently, the lack of good bone segmentation leads to inaccurate attenuation coefficient map which produces a strong spatial bias of the PET activity. For instance, ignoring the bone attenuation coefficients in the head can lead to 20% underestimation of PET activity [107].
Another challenge is the assignment of attenuation coefficients to each tissue type. Currently, there is no agreement on the value for each class where. For instance, soft tissue linear attenuation coefficient ranges between 0.094 and 0.100, while trabecular bone has one single value assigned to 0.110 and cortical bone ranges from 0.120 to 0.172 [47]. This variation of attenuation coefficients effects the accuracy of attenuation correction map even if the segmentation accuracy is high.
One more important limitation is the assignment of discrete attenuation coefficients while the density of body tissue is represented by continuous values. There is a need to explore and develop segmentation methods that measure continuous attenuation coefficient values for bone and other tissues in order to obtain accurate PET quantification.
Moreover, there is an interpatient variability of tissue attenuation coefficient based on gender and age. This variability can cause non-negligible errors especially in the tissue regions that show high interpatient variability such as bone.
Additionally, there are other, clinically related, challenges and difficulties that affect MR image-based attenuation correction which are outside the scope of this review such as body truncation artifacts, the presence of ancillary objects during the scanning such as the patient bed, MR coils, positioning aids, and medical probes [108].
Emerging Techniques
Deep learning-based methods were proposed for pelvic and prostate PET attenuation correction. For instance, Bradshaw et al. [109] applied a 3D CNN called DeepMedic [110] to segment pelvis T1- and T2-weighted MR images for PET attenuation correction. Beside segmentation-based methods, there are other works that applied deep learning for attenuation correction for brain [111] and pelvic [112] to learn the relationship between MR and CT images then generate pseudo CT images. Deep learning showed its superiority to classical techniques for MR images segmentation. However, the applications of deep learning for brain MR image segmentation for attenuation correction are limited.
There are other studies that utilized deep learning-based methods for MR image-guided radiation therapy and treatment planning for brain tumor [113,114,115], prostate/pelvic region [116] and other different whole-body tissues [117]. These studies applied different deep models such as standard CNN, dilated CNN, and generative adversarial networks (GAN). A comparison study [113] between one segmentation, four atlas, and one deep learning methods to evaluate the MR image-based radiotherapy planning in the pelvic region study showed the outperformance of deep learning method in terms of segmentation accuracy, CT generation accuracy, and dosimetric evaluation.
In terms of MR sequences, currently there is room for improvement of the PET attenuation correction performance by using more sophisticated MR sequences with high signal intensity for bone such as UTE, ZTE, or Dixon sequences. These sequences are able to capture very short T2 values [57, 60] which enables accurate detection of bone tissue and, in turn, improves the attenuation correction map. Another elegant way is acquiring UTE sequences at different echo time to extract more information. Moreover, Dixon sequences provides easy access to four tissue classes which are soft tissue, air, fat, and lung.
The usage of the special sequences such as UTE, ZTE, and Dixon images along with robust segmentation techniques and continuous linear attenuation coefficients can achieve better accuracy for PET attenuation correction than atlas-based methods. However, the major drawback of this sequences in the long acquisition time potentially hampering clinical flow.
Conclusion
This article presented the application of machine learning techniques for MR image segmentation-based PET attenuation correction in hybrid PET/MR scanners. Among machine learning techniques, clustering, classification, and deep learning are proposed in the literature for tissue segmentation. Deep learning approach outperforms other classical machine learning techniques in this task. Although much progress has been made recently with machine learning methods for segmentation, the reported deep learning methods are fewer especially for brain PET attenuation correction. In summary, deep learning is needed to improve the segmentation and attenuation correction accuracy and intensive clinical evaluation studies of deep learning approach are required.
References
Liu F, Jang H, Kijowski R, Zhao G, Bradshaw T, McMillan AB: A deep learning approach for 18f-FDG PET attenuation correction. EJNMMI Physics 5 (1): 24, 2018. https://doi.org/10.1186/s40658-018-0225-8
Cabello J, Lukas M, Förster S, Pyka T, Nekolla SG, Ziegler SI, et al.: Mr-based attenuation correction using ultrashort-echo-time pulse sequences in dementia patients. J Nucl Med 56(3):423–429, 2015
Aitken AP, Giese D, Tsoumpas C, Schleyer P, Kozerke S, Prieto C, Schaeffter T: Improved ute-based attenuation correction for cranial pet-mr using dynamic magnetic field monitoring. Medical physics 41(1):012302, 2014
Shi K, Fürst S, Sun L, Lukas M, Navab N, Förster S, Ziegler SI: Individual refinement of attenuation correction maps for hybrid pet/mr based on multi-resolution regional learning. Computerized Medical Imaging and Graphics 60: 50–57, 2017
Berker Y, Franke J, Salomon A, Palmowski M, Donker HCW, Temur Y, Mottaghy FM, Kuhl C, Izquierdo-Garcia D, Fayad ZA, Kiessling F, Schulz V: MRI-based attenuation correction for hybrid PET/MRI Systems: A 4-class tissue segmentation technique using a combined ultrashort-echo-time/Dixon MRI sequence. Journal of Nuclear Medicine 53(5):796–804, 2012. https://doi.org/10.2967/jnumed.111.092577
Izquierdo-Garcia D, Hansen AE, Förster S, Benoit D, Schachoff S, Fürst S, Chen KT, Chonde DB, Catana C: An SPM8-Based Approach for Attenuation Correction Combining Segmentation and rigid Template Formation: Application to Simultaneous PET/MR Brain Imaging. Journal of Nuclear Medicine 55 (11): 1825–1830, 2014. https://doi.org/10.2967/jnumed.113.136341
Bammer R: Basic principles of diffusion-weighted imaging. European journal of radiology 45(3):169–184, 2003
Khateri P, Saligheh Rad H, Jafari AH, Fathi Kazerooni A, Akbarzadeh A, Shojae Moghadam M, Aryan A, Ghafarian P, Ay MR: Generation of a Four-Class Attenuation Map for MRI-Based Attenuation Correction of PET Data in the Head Area Using a Novel Combination of STE/Dixon-MRI and FCM Clustering. Molecular Imaging and Biology 17(6):884–892, 2015. https://doi.org/10.1007/s11307-015-0849-1
Waldman A, Rees JH, Brock CS, Robson MD, Gatehouse PD, Bydder GM: Mri of the brain with ultra-short echo-time pulse sequences. Neuroradiology 45(12):887–892, 2003
Weiger M, Pruessmann KP (2012) Mri with zero echo time. eMagRes
Hylton N: Dynamic contrast-enhanced magnetic resonance imaging as an imaging biomarker. J Clin Oncol 24(20):3293–3298, 2006
Mugler III JP, Brookeman JR: Three-dimensional magnetization-prepared rapid gradient-echo imaging (3d mp rage). Magnetic Resonance in Medicine 15(1):152–157, 1990
Pustina D, Coslett HB, Turkeltaub PE, Tustison N, Schwartz MF, Avants B: Automated segmentation of chronic stroke lesions using LINDA: Lesion identification with neighborhood data analysis. Human Brain Mapping 37(4):1405–1421, 2016. https://doi.org/10.1002/hbm.23110
Praveen GB, Agrawal A, Sundaram P, Sardesai S: Ischemic stroke lesion segmentation using stacked sparse autoencoder. Computers in Biology and Medicine 99: 38–52, 2018. https://doi.org/10.1016/j.compbiomed.2018.05.027
Boldsen JK, Engedal TS, Pedraza S, Cho T-H, Thomalla G, Nighoghossian N, Baron J-C, Fiehler J, Østergaard L, Mouridsen K: Better Diffusion Segmentation in Acute Ischemic Stroke Through Automatic Tree Learning Anomaly Segmentation. Frontiers in Neuroinformatics 12:21, 2018. https://doi.org/10.3389/fninf.2018.00021
Salem M, Cabezas M, Valverde S, Pareto D, Oliver A, Salvi J, Rovira, Lladó X: A supervised framework with intensity subtraction and deformation field features for the detection of new T2-w lesions in multiple sclerosis. NeuroImage: Clinical 17:607–615, 2018. https://doi.org/10.1016/j.nicl.2017.11.015
Roy S, He Q, Sweeney E, Carass A, Reich DS, Prince JL, Pham DL: Subject-Specific Sparse Dictionary Learning for Atlas-Based Brain MRI Segmentation. IEEE Journal of Biomedical and Health Informatics 19 (5): 1598–1609, 2015. https://doi.org/10.1109/JBHI.2015.2439242
Brosch T, Yoo Y, Tang LisaYW, Li DKB, Traboulsee A, Tam R: Deep convolutional encoder networks for multiple sclerosis lesion segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2015, pp 3–11
Brosch T, Tang LYW, Yoo Y, Li DKB, Traboulsee A, Tam R: Deep 3d Convolutional Encoder Networks With Shortcuts for Multiscale Feature Integration Applied to Multiple Sclerosis Lesion Segmentation. IEEE Transactions on Medical Imaging 35 (5): 1229–1239, 2016. https://doi.org/10.1109/TMI.2016.2528821
van Opbroek A, Achterberg HC, de Bruijne M: Feature-Space Transformation Improves Supervised Segmentation Across Scanners. In: (Bhatia K, Lombaert H, Eds.) Machine Learning Meets Medical Imaging. Lecture Notes in Computer Science. Springer International Publishing, 2015, pp 85–93
Liu M, Cheng D, Wang K, Wang Y, Initiative ADN, et al: Multi-modality cascaded convolutional neural networks for alzheimer’s disease diagnosis. Neuroinformatics 16(3-4):295–308, 2018
Rundo L, Militello C, Tangherloni A, Russo G, Vitabile S, Gilardi MC, Mauri G: NeXt for neuro-radiosurgery: A fully automatic approach for necrosis extraction in brain tumor MRI using an unsupervised machine learning technique. International Journal of Imaging Systems and Technology 28(1):21–37, 2018. https://doi.org/10.1002/ima.22253
Polly FP, Shil SK, Hossain MA, Ayman A, Jang YM: Detection and classification of HGG and LGG brain tumor using machine learning. In: 2018 International Conference on Information Networking (ICOIN), 2018, pp 813–817
Osman AFI: Automated Brain Tumor Segmentation on Magnetic Resonance Images and Patient’s Overall Survival Prediction Using Support Vector Machines. In: (Crimi A, Bakas S, Kuijf H, Menze B, Reyes M, Eds.) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Lecture Notes in Computer Science. Springer International Publishing, 2018, pp 435–449
Prior FW, Fouke SJ, Benzinger T, Boyd A, Chicoine M, Cholleti S, Kelsey M, Keogh B, Kim L, Milchenko M, Politte DG, Tyree S, Weinberger K, Marcus D: Predicting a multi-parametric probability map of active tumor extent using random forests. In: 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2013, pp 6478–6481
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin P-M, Larochelle H: Brain tumor segmentation with Deep Neural Networks. Medical Image Analysis 35:18–31, 2017. https://doi.org/10.1016/j.media.2016.05.004
Sedlar S: Brain Tumor Segmentation Using a Multi-path CNN Based Method. In: (Crimi A, Bakas S, Kuijf H, Menze B, Reyes M, Eds.) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Lecture Notes in Computer Science. Springer International Publishing, 2018, pp 403–422
Soltaninejad M, Yang G, Lambrou T, Allinson N, Jones TL, Barrick TR, Howe FA, Ye X: Supervised learning based multimodal MRI brain tumour segmentation using texture features from supervoxels. Computer Methods and Programs in Biomedicine 157:69–84, 2018. https://doi.org/10.1016/j.cmpb.2018.01.003
Stone JR, Wilde EA, Taylor BA, Tate DF, Levin H, Bigler ED, Scheibel RS, Newsome MR, Mayer AR, Abildskov T, Black GM, Lennon MJ, York GE, Agarwal R, DeVillasante J, Ritter JL, Walker PB, Ahlers ST, Tustison NJ: Supervised learning technique for the automated identification of white matter hyperintensities in traumatic brain injury. Brain Injury 30(12):1458–1468, 2016. https://doi.org/10.1080/02699052.2016.1222080
Wu Z, Gao Y, Shi F, Jewells V, Shen D: Automatic Hippocampal Subfield Segmentation from 3t Multi-modality Images. In: (Wang L, Adeli E, Wang Q, Shi Y, Suk H-I, Eds.) Machine Learning in Medical Imaging. Lecture Notes in Computer Science. Springer International Publishing, 2016, pp 229–236
Bonte S, Goethals I, Van Holen R: Machine learning based brain tumour segmentation on limited data using local texture and abnormality. Computers in Biology and Medicine 98:39–47, 2018. https://doi.org/10.1016/j.compbiomed.2018.05.005
Rincón M, Díaz-López E, Selnes P, Vegge K, Altmann M, Fladby T, Børnerud A: Improved Automatic Segmentation of White Matter Hyperintensities in MRI Based on Multilevel Lesion Features. Neuroinformatics 15(3):231–245, 2017. https://doi.org/10.1007/s12021-017-9328-y
Magome T, Arimura H, Kakeda S, Yamamoto D, Kawata Y, Yamashita Y, Higashida Y, Toyofuku F, Ohki M, Korogi Y: Automated segmentation method of white matter and gray matter regions with multiple sclerosis lesions in MR images. Radiological Physics and Technology 4(1):61–72, 2011. https://doi.org/10.1007/s12194-010-0106-x
Fan W, Hu C, Du J, Bouguila N: A Novel Model-Based Approach for Medical Image Segmentation Using Spatially Constrained Inverted Dirichlet Mixture Models. Neural Processing Letters 47(2):619–639, 2018. https://doi.org/10.1007/s11063-017-9672-9
Ahmadvand A, Daliri MR, Zahiri SM: Segmentation of brain MR images using a proper combination of DCS based method with MRF. Multimedia Tools and Applications 77(7):8001–8018, 2018. https://doi.org/10.1007/s11042-017-4696-8
Elliott C, Arnold DL, Collins DL, Arbel T: A generative model for automatic detection of resolving multiple sclerosis lesions. In: Bayesian and grAphical Models for Biomedical Imaging. Springer, 2014, pp 118–129
Tamajka M, Benesova W: Automatic brain segmentation method based on supervoxels. In: 2016 International Conference on Systems, Signals and Image Processing (IWSSIP), 2016 , pp 1–4
De A, Guo C: An image segmentation method based on the fusion of vector quantization and edge detection with applications to medical image processing. International Journal of Machine Learning and Cybernetics 5(4):543–551, 2014. https://doi.org/10.1007/s13042-013-0205-1
Kothavari K, Arunadevi B, Deepa SN: A Hybrid DE-RGSO-ELM for Brain Tumor Tissue Categorization in 3d Magnetic Resonance Images. Mathematical Problems in Engineering 2014:1–14, 2014. https://doi.org/10.1155/2014/291581
Yilmaz B, Durdu A, Emlik GD: A new method for skull stripping in brain MRI using multistable cellular neural networks. Neural Computing and Applications 29(8):79–95, 2018. https://doi.org/10.1007/s00521-016-2834-2
Mekhmoukh A, Mokrani K: Improved Fuzzy C-Means based Particle Swarm Optimization (PSO) initialization and outlier rejection with level set methods for MR brain image segmentation. Computer Methods and Programs in Biomedicine 122(2):266–281, 2015. https://doi.org/10.1016/j.cmpb.2015.08.001
Xie Y, Tao X: White matter lesion segmentation using machine learning and weakly labeled MR images. In: Medical Imaging 2011: Image Processing, vol 7962. International Society for Optics and Photonics, 2011, p 79622G
Rajchl M, Lee MCH, Oktay O, Kamnitsas K, Passerat-Palmbach J, Bai W, Damodaram M, Rutherford MA, Hajnal JV, Kainz B, Rueckert D: DeepCut: Object Segmentation From Bounding Box Annotations Using Convolutional Neural Networks. IEEE Transactions on Medical Imaging 36(2):674–683, 2017. https://doi.org/10.1109/TMI.2016.2621185
Pedemonte S, Bizzo B, Pomerantz S, Tenenholtz N, Wright B, Walters M, Doyle S, McCarthy A, De Almeida RR, Andriole K, Michalski M, Gilberto Gonzalez R: Detection and Delineation of Acute Cerebral Infarct on DWI Using Weakly Supervised Machine Learning. In: (Frangi AF, Schnabel JA, Davatzikos C, Alberola-López C, Fichtinger G, Eds.) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Lecture Notes in Computer Science. Springer International Publishing, 2018, pp 81–88
Valverde S, Cabezas M, Roura E, González-Villà? S, Pareto D, Vilanova JC, Ramió-Torrentà? L, Rovira l, Oliver A, Lladó X: Improving automated multiple sclerosis lesion segmentation with a cascaded 3d convolutional neural network approach. NeuroImage 155:159–168, 2017. https://doi.org/10.1016/j.neuroimage.2017.04.034
Sjölund J, Forsberg D, Andersson M, Knutsson H: Generating patient specific pseudo-CT of the head from mr using atlas-based regression. Physics in Medicine & Biology 60(2):825, 2015
Mehranian A, Arabi H, Zaidi H: Vision 20/20: Magnetic resonance imaging-guided attenuation correction in PET/MRI: Challenges, solutions, and opportunities. Medical Physics 43(3):1130–1155, 2016. https://doi.org/10.1118/1.4941014
Wagenknecht G, Kaiser H-J, Mottaghy FM, Herzog H: MRI for attenuation correction in PET: methods and challenges. Magnetic Resonance Materials in Physics, Biology and Medicine 26(1):99–113, 2013. https://doi.org/10.1007/s10334-012-0353-4
Chen Z, Jamadar SD, Li S, Sforazzini F, Baran J, Ferris N, Shah NJ, Egan GF: From simultaneous to synergistic mr-pet brain imaging: A review of hybrid mr-pet imaging methodologies. Human brain mapping 39(12):5126–5144, 2018
Ladefoged CN, Benoit D, Law I, Holm S, Kær A, Højgaard L, Hansen AE, Andersen FL: Region specific optimization of continuous linear attenuation coefficients based on UTE (RESOLUTE): application to PET/MR brain imaging. Physics in Medicine & Biology 60(20):8047, 2015. https://doi.org/10.1088/0031-9155/60/20/8047
(2018) X-ray mass attenuation coefficients. https://physics.nist.gov/PhysRefData/XrayMassCoef/tab2.html [Online; accessed 26-February-2019]
Kazerooni AF, Ay MR, Arfaie S, Khateri P, Rad HS: Single STE-MR Acquisition in MR-Based Attenuation Correction of Brain PET Imaging Employing a Fully Automated and Reproducible Level-Set Segmentation Approach. Molecular Imaging and Biology 19(1):143–152, 2017. https://doi.org/10.1007/s11307-016-0990-5
An HJ, Seo S, Kang H, Choi H, Cheon GJ, Kim H-J, Lee DS, Song IC, Kim YK, Lee JS: MRI-Based Attenuation Correction for PET/MRI Using Multiphase Level-Set Method. Journal of Nuclear Medicine 57(4):587–593, 2016. https://doi.org/10.2967/jnumed.115.163550
Catana C, Kouwe A, Benner T, Michel CJ, Hamm M, Fenchel M, Fischl B, Rosen B, Schmand M, Sorensen AG: Toward Implementing an MRI-Based PET Attenuation-Correction Method for Neurologic Studies on the MR-PET Brain Prototype. Journal of Nuclear Medicine 51(9):1431–1438, 2010. https://doi.org/10.2967/jnumed.109.069112
Delso G, Zeimpekis K, Carl M, Wiesinger F, Hüllner M, Veit-Haibach P: Cluster-based segmentation of dual-echo ultra-short echo time images for PET/MR bone localization. EJNMMI Physics 1(1):7, 2014. https://doi.org/10.1186/2197-7364-1-7
Jang H, Liu F, Bradshaw T, McMillan AB: Rapid dual-echo ramped hybrid encoding MR-based attenuation correction (dRHE-MRAC) for PET/MR. Magnetic Resonance in Medicine 79(6):2912–2922, 2018. https://doi.org/10.1002/mrm.26953
Khalifé M, Fernandez B, Jaubert O, Soussan M, Brulon V, Buvat I, Claude Comtat: Subject-specific bone attenuation correction for brain PET/MR: can ZTE-MRI substitute CT scan accurately? Physics in Medicine & Biology 62(19):7814, 2017. https://doi.org/10.1088/1361-6560/aa8851
Juttukonda MR, Mersereau BG, Chen Y, Su Y, Rubin BG, Benzinger TLS, Lalush DS, An H: MR-based attenuation correction for PET/MRI neurological studies with continuous-valued attenuation coefficients for bone through a conversion from R2* to CT-Hounsfield units. NeuroImage 112:160–168, 2015. https://doi.org/10.1016/j.neuroimage.2015.03.009
Yang X, Fei B: Multiscale segmentation of the skull in MR images for MRI-based attenuation correction of combined MR/PET. Journal of the American Medical Informatics Association 20(6):1037–1045, 2013. https://doi.org/10.1136/amiajnl-2012-001544
Keereman V, Fierens Y, Broux T, Deene YD, Lonneux M, Vandenberghe S: MRI-Based Attenuation Correction for PET/MRI Using Ultrashort Echo Time Sequences. Journal of Nuclear Medicine 51(5):812–818, 2010. https://doi.org/10.2967/jnumed.109.065425
Santos Ribeiro A, Rota Kops E, Herzog H, Almeida P: Skull segmentation of UTE MR images by probabilistic neural network for attenuation correction in PET/MR. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 702:114–116, 2013. https://doi.org/10.1016/j.nima.2012.09.005
Khateri P, Rad HS, Jafari AH, Ay MR: A novel segmentation approach for implementation of MRAC in head PET/MRI employing Short-TE MRI and 2-point Dixon method in a fuzzy C-means framework. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 734:171–174, 2014. https://doi.org/10.1016/j.nima.2013.09.006
Fei B, Yang X, Nye JA, Aarsvold JN, Raghunath N, Cervo M, Stark R, Meltzer CC, Votaw JR: MR/PET quantification tools: Registration, segmentation, classification, and MR-based attenuation correction. Medical Physics 39(10):6443–6454, 2012. https://doi.org/10.1118/1.4754796
Chan SLS, Jeffree RL, Fay M, Crozier S, Yang Z, Gal Y, Thomas P: Automated Classification of Bone and Air Volumes for Hybrid PET-MRI Brain Imaging. In: 2013 International Conference on Digital Image Computing: Techniques and Applications (DICTA), 2013, pp 1–8
Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB: Deep learning mr imaging–based attenuation correction for pet/mr imaging. Radiology 286(2):676–684, 2017
Jang H, Liu F, Zhao G, Bradshaw T, McMillan AB: Technical Note: Deep learning based MRAC using rapid ultrashort echo time imaging. Medical Physics 45 (8): 3697–3704, 2018. https://doi.org/10.1002/mp.12964
Gong K, Yang J, Kim K, El Fakhri G, Seo Y, Li Q: Attenuation correction for brain pet imaging using deep neural network based on dixon and zte mr images. Physics in medicine and biology 63.12:125011, 2018
Burgos N, Cardoso MJ, Thielemans K, Modat M, Pedemonte S, Dickson J, Barnes A, Ahmed R, Mahoney CJ, Schott JM, et al: Attenuation correction synthesis for hybrid pet-mr scanners: application to brain studies. IEEE Trans. Med. Imaging 33(12):2332–2341, 2014
Schreibmann E, Nye JA, Schuster DM, Martin DR, Votaw J, Fox T: Mr-based attenuation correction for hybrid pet-mr brain imaging systems using deformable image registration. Medical physics 37(5):2101–2109, 2010
Mérida I, Costes N, Heckemann RA, Drzezga A, Förster S, Hammers A: Evaluation of several multi-atlas methods for pseudo-ct generation in brain mri-pet attenuation correction. In: Biomedical Imaging (ISBI), 2015 IEEE 12th International Symposium on, IEEE, 2015, pp 1431–1434
Chen KT, Izquierdo-Garcia D, Poynton CB, Chonde DB, Catana C: On the accuracy and reproducibility of a novel probabilistic atlas-based generation for calculation of head attenuation maps on integrated pet/mr scanners. European journal of nuclear medicine and molecular imaging 44(3):398–407, 2017
Su K-H, Hu L, Stehning C, Helle M, Qian P, Thompson CL, Pereira GC, Jordan DW, Herrmann KA, Traughber M, Muzic RF, Traughber BJ: Generation of brain pseudo-CTs using an undersampled, single-acquisition UTE-mDixon pulse sequence and unsupervised clustering. Medical Physics 42(8):4974–4986, 2015. https://doi.org/10.1118/1.4926756
Koesters T, Friedman KP, Fenchel M, Zhan Y, Hermosillo G, Babb J, Jelescu IO, Faul D, Boada FE, Shepherd TM: Dixon Sequence with Superimposed Model-Based Bone Compartment Provides Highly Accurate PET/MR Attenuation Correction of the Brain. Journal of Nuclear Medicine 57 (6): 918–924, 2016. https://doi.org/10.2967/jnumed.115.166967
Xiang L, Wang Q, Jin X, Nie D, Qiao Y, Shen D (2017) Deep Embedding Convolutional Neural Network for Synthesizing CT Image from T1-Weighted MR Image. arXiv:1709.02073
Wu Y, Yang W, Lu L, Lu Z, Zhong L, Huang M, Feng Y, Feng Q, Chen W: Prediction of CT Substitutes from MR Images Based on Local Diffeomorphic Mapping for Brain PET Attenuation Correction. Journal of Nuclear Medicine 57 (10): 1635–1641, 2016. https://doi.org/10.2967/jnumed.115.163121
Wu Y, Yang W, Lu L, Lu Z, Zhong L, Yang R, Huang M, Feng Y, Chen W, Feng Q: Prediction of ct substitutes from mr images based on local sparse correspondence combination. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, 2015, pp 93–100
Johansson A, Karlsson M, Nyholm T: CT substitute derived from MRI sequences with ultrashort echo time. Medical physics 38 (5): 2708–2714, 2011
Zhong L, Lin L, Lu Z, Wu Y, Lu Z, Huang M, Yang W, Feng Q: Predict CT image from MRI data using KNN-regression with learned local descriptors, 2016, pp 743–746
Yang W, Zhong L, Chen Y, Lin L, Lu Z, Liu S, Wu Y, Feng Q, Chen W: Predicting CT Image From MRI Data Through Feature Matching With Learned Nonlinear Local Descriptors. IEEE Transactions on Medical Imaging 37(4):977–987, 2018. https://doi.org/10.1109/TMI.2018.2790962
Yang X, Lei Y, Shu H-K, Rossi P, Mao H, Shim H, Curran WJ, Liu T: Pseudo CT estimation from MRI using patch-based random forest. In: Medical Imaging 2017: Image Processing, vol 10133. International Society for Optics and Photonics, 2017, p 101332Q
Huynh T, Gao Y, Kang J, Wang L, Zhang P, Lian J, Shen D: Estimating CT Image From MRI Data Using Structured Random Forest and Auto-Context Model. IEEE Transactions on Medical Imaging 35(1):174–183, 2016. https://doi.org/10.1109/TMI.2015.2461533
Torrado-Carvajal A, Herraiz JL, Alcain E, Montemayor AS, Garcia-Cañamaque L, Hernandez-Tamames JA, Rozenholc Y, Malpica N: Fast Patch-Based Pseudo-CT Synthesis from T1-Weighted MR Images for PET/MR Attenuation Correction in Brain Studies. Journal of Nuclear Medicine 57(1):136–143, 2016. https://doi.org/10.2967/jnumed.115.156299
Roy S, Wang W-T, Carass A, Prince JL, Butman JA, Pham DL: PET Attenuation Correction Using Synthetic CT from Ultrashort Echo-Time MR Imaging. Journal of Nuclear Medicine 55(12):2071–2077, 2014. https://doi.org/10.2967/jnumed.114.143958
Arabi H, Zeng G, Zheng G, Zaidi H: Novel adversarial semantic structure deep learning for mri-guided attenuation correction in brain pet/mri. European journal of nuclear medicine and molecular imaging 46(13):2746–2759, 2019
Badrinarayanan V, Kendall A, Cipolla R (2015) Segnet: A deep convolutional encoder-decoder architecture for image segmentation. arXiv:1511.00561
Liu F, Jang H, Kijowski R, Zhao G, Bradshaw T, McMillan AB: A deep learning approach for 18 f-fdg pet attenuation correction. EJNMMI physics 5(1):24, 2018
Taha AA, Hanbury A: Metrics for evaluating 3d medical image segmentation: analysis, selection, and tool. BMC medical imaging 15(1):29, 2015
Aasheim LB, Karlberg A, Goa PE, Håberg A, Sørhaug S, Fagerli U-M, Eikenes L: PET/MR brain imaging: evaluation of clinical UTE-based attenuation correction. European Journal of Nuclear Medicine and Molecular Imaging 42(9):1439–1446, 2015. https://doi.org/10.1007/s00259-015-3060-3
Choi H, Cheon GJ, Kim H-J, Choi SH, Lee JS, Kim Y, Kang KW, Chung JK, Kim EE, Lee DS: Segmentation-Based MR Attenuation Correction Including Bones Also Affects Quantitation in Brain Studies: An Initial Result of 18f-FP-CIT PET/MR for Patients with Parkinsonism. Journal of Nuclear Medicine 55(10):1617–1622, 2014. https://doi.org/10.2967/jnumed.114.138636
Delso G, Wiesinger F, Sacolick LI, Kaushik SS, Shanbhag DD, Hllner M, Veit-Haibach P: Clinical Evaluation of Zero-Echo-Time MR Imaging for the Segmentation of the Skull. Journal of Nuclear Medicine 56(3):417–422, 2015. https://doi.org/10.2967/jnumed.114.149997
Sekine T, Voert EEGW, Warnock G, Buck A, Huellner M, Veit-Haibach P, Delso G: Clinical Evaluation of Zero-Echo-Time Attenuation Correction for Brain 18f-FDG PET/MRI: Comparison with Atlas Attenuation Correction. Journal of Nuclear Medicine 57(12):1927–1932, 2016. https://doi.org/10.2967/jnumed.116.175398
Anazodo UC, Thiessen JD, Ssali T, Mandel J, Günther M, Butler J, Pavlosky W, Prato FS, Thompson RT, Lawrence S, S K: Feasibility of simultaneous whole-brain imaging on an integrated PET-MRI system using an enhanced 2-point Dixon attenuation correction method. Frontiers in Neuroscience 8:434, 2015. https://doi.org/10.3389/fnins.2014.00434
Andersen FL, Ladefoged CN, Beyer T, Keller SH, Hansen AE, Højgaard L, Kjær A, Law I, Holm S: Combined PET/MR imaging in neurology: MR-based attenuation correction implies a strong spatial bias when ignoring bone. NeuroImage 84:206–216, 2014. https://doi.org/10.1016/j.neuroimage.2013.08.042
Dickson JC, O’Meara C, Barnes A: A comparison of CT- and MR-based attenuation correction in neurological PET. European Journal of Nuclear Medicine and Molecular Imaging 41(6):1176–1189, 2014. https://doi.org/10.1007/s00259-013-2652-z
Baran J, Chen Z, Sforazzini F, Ferris N, Jamadar S, Schmitt B, Faul D, Shah NJ, Cholewa M, Egan GF: Accurate hybrid template–based and mr-based attenuation correction using ute images for simultaneous pet/mr brain imaging applications. BMC medical imaging 18(1):41, 2018
Ladefoged CN, Law I, Anazodo U, Lawrence KS, Izquierdo-Garcia D, Catana C, Burgos N, Cardoso MJ, Ourselin S, Hutton B, et al: A multi-centre evaluation of eleven clinically feasible brain pet/mri attenuation correction techniques using a large cohort of patients. Neuroimage 147:346–359, 2017
Izquierdo-Garcia D, Hansen AE, Förster S, Benoit D, Schachoff S, Fürst S, Chen KT, Chonde DB, Catana C: An spm8-based approach for attenuation correction combining segmentation and nonrigid template formation: application to simultaneous pet/mr brain imaging. Journal of Nuclear Medicine 55(11):1825–1830, 2014
Zaini MHM, Shapiai MI, Mohamed AR, Fauzi H, Ibrahim Z, Adam A: Hippocampal segmentation using structured extreme learning machine with bag of features. In: 2017 International Conference on Robotics, Automation and Sciences (ICORAS), 2017, pp 1–5
Ayerdi B, Savio A, Graña M: Meta-ensembles of classifiers for alzheimer’s disease detection using independent roi features. In: International Work-Conference on the Interplay Between Natural and Artificial Computation. Springer, 2013, pp 122–130
Rachmadi MF, Valdés-Hernández MC, Agan MLF, Di Perri C, Komura T: Segmentation of white matter hyperintensities using convolutional neural networks with global spatial information in routine clinical brain MRI with none or mild vascular pathology. Computerized Medical Imaging and Graphics 66:28–43, 2018. https://doi.org/10.1016/j.compmedimag.2018.02.002
Roy PK, Bhuiyan A, Janke A, Desmond PM, Wong TY, Abhayaratna WP, Storey E, Ramamohanarao K: Automatic white matter lesion segmentation using contrast enhanced FLAIR intensity and Markov Random Field. Computerized Medical Imaging and Graphics 45:102–111, 2015. https://doi.org/10.1016/j.compmedimag.2015.08.005
Serag A, Boardman JP, Wilkinson AG, Macnaught G, Semple SI: A sparsity-based atlas selection technique for multiple-atlas segmentation: Application to neonatal brain labeling. In: 2016 24th Signal Processing and Communication Application Conference (SIU), 2016, pp 2265–2268
Amin J, Sharif M, Raza M, Yasmin M (2018) Detection of Brain Tumor based on Features Fusion and Machine Learning. Journal of Ambient Intelligence and Humanized Computing. pp 1–17
Valverde S, Cabezas M, Roura E, González-Villà S, Pareto D, Vilanova JC, Ramió-Torrentà L, Rovira A, Oliver A, Lladó X: Improving automated multiple sclerosis lesion segmentation with a cascaded 3d convolutional neural network approach. NeuroImage 155:159–168, 2017
Sudre CH, Li W, Vercauteren T, Ourselin S, Cardoso MJ: Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, Springer, pp 240–248, 2017
Salehi SSM, Erdogmus D, Gholipour A: Tversky loss function for image segmentation using 3d fully convolutional deep networks. In: International Workshop on Machine Learning in Medical Imaging. Springer, 2017, pp 379–387
Chen Y, An H: Attenuation Correction of PET/MR Imaging. Magnetic Resonance Imaging Clinics of North America 25(2):245–255, 2017. https://doi.org/10.1016/j.mric.2016.12.001
Bezrukov I, Mantlik F, Schmidt H, Schölkopf B, Pichler BJ: MR-Based PET Attenuation Correction for PET/MR Imaging. Seminars in Nuclear Medicine 43(1):45–59, 2013. https://doi.org/10.1053/j.semnuclmed.2012.08.002
Bradshaw TJ, Zhao G, Jang H, Liu F, McMillan AB: Feasibility of Deep Learning–Based PET/MR Attenuation Correction in the Pelvis Using Only Diagnostic MR Images. Tomography 4(3):138–147, 2018. https://doi.org/10.18383/j.tom.2018.00016
Kamnitsas K, Ferrante E, Parisot S, Ledig C, Nori AV, Criminisi A, Rueckert D, Glocker B: DeepMedic for Brain Tumor Segmentation. In: (Crimi A, Menze B, Maier O, Reyes M, Winzeck S, Handels H, Eds.) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Lecture Notes in Computer Science. Springer International Publishing, 2016, pp 138–149
Roy S, Butman JA, Pham DL: Synthesizing ct from ultrashort echo-time mr images via convolutional neural networks. In: International Workshop on Simulation and Synthesis in Medical Imaging. Springer, 2017, pp 24–32
Leynes AP, Yang J, Wiesinger F, Kaushik SS, Shanbhag DD, Seo Y, Hope TA, Larson PEZ: Zero-Echo-Time and Dixon Deep Pseudo-CT (ZeDD CT): Direct Generation of Pseudo-CT Images for Pelvic PET/MRI Attenuation Correction Using Deep Convolutional Neural Networks with Multiparametric MRI. Journal of Nuclear Medicine 59(5):852–858, 2018. https://doi.org/10.2967/jnumed.117.198051
Han X: Mr-based synthetic ct generation using a deep convolutional neural network method. Medical physics 44(4):1408–1419, 2017
Emami H, Dong M, Nejad-Davarani SP, Glide-Hurst CK: Generating synthetic CTs from magnetic resonance images using generative adversarial networks. Medical Physics 45(8):3627–3636, 2018. https://doi.org/10.1002/mp.13047
Dinkla AM, Wolterink JM, Maspero M, Savenije MHF, Verhoeff JJC, Seravalli E, Išgum I, Seevinck PR, van den Berg CAT: MR-Only Brain Radiation Therapy: Dosimetric Evaluation of Synthetic CTs Generated by a Dilated Convolutional Neural Network. International Journal of Radiation Oncology*Biology*Physics 102(4):801–812, 2018. https://doi.org/10.1016/j.ijrobp.2018.05.058
Chen S, Qin A, Zhou D, Yan D: Technical Note: U-net-generated synthetic CT images for magnetic resonance imaging-only prostate intensity-modulated radiation therapy treatment planning. Medical Physics 45(12):5659–5665, 2018. https://doi.org/10.1002/mp.13247
Fu Y, Mazur TR, Wu X, Liu S, Chang X, Lu Y, Li HH, Kim H, Roach MC, Henke L, Yang D: A novel MRI segmentation method using CNN-based correction network for MRI-guided adaptive radiotherapy. Medical Physics 45(11):5129–5137, 2018. https://doi.org/10.1002/mp.13221
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mecheter, I., Alic, L., Abbod, M. et al. MR Image-Based Attenuation Correction of Brain PET Imaging: Review of Literature on Machine Learning Approaches for Segmentation. J Digit Imaging 33, 1224–1241 (2020). https://doi.org/10.1007/s10278-020-00361-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-020-00361-x