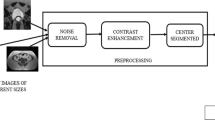
Abstract
Transfer learning using deep pre-trained convolutional neural networks is increasingly used to solve a large number of problems in the medical field. In spite of being trained using images with entirely different domain, these networks are flexible to adapt to solve a problem in a different domain too. Transfer learning involves fine-tuning a pre-trained network with optimal values of hyperparameters such as learning rate, batch size, and number of training epochs. The process of training the network identifies the relevant features for solving a specific problem. Adapting the pre-trained network to solve a different problem requires fine-tuning until relevant features are obtained. This is facilitated through the use of large number of filters present in the convolutional layers of pre-trained network. A very few features out of these features are useful for solving the problem in a different domain, while others are irrelevant, use of which may only reduce the efficacy of the network. However, by minimizing the number of filters required to solve the problem, the efficiency of the training the network can be improved. In this study, we consider identification of relevant filters using the pre-trained networks namely AlexNet and VGG-16 net to detect cervical cancer from cervix images. This paper presents a novel hybrid transfer learning technique, in which a CNN is built and trained from scratch, with initial weights of only those filters which were identified as relevant using AlexNet and VGG-16 net. This study used 2198 cervix images with 1090 belonging to negative class and 1108 to positive class. Our experiment using hybrid transfer learning achieved an accuracy of 91.46%.

















Similar content being viewed by others
References
J. Ferlay, I. Soerjomataram, M. Ervik, R. Dikshit, S. Eser, C. Mathers, M. Rebelo, D. M. Parkin, D. Forman and F. Bray 2013 GLOBOCAN 2012 v1.0, Cancer incidence and mortality worldwide: IARC CancerBase No. 11, Lyon, France: IARC [internet]. Available from:http://globocan.iarc.fr, . Last accessed on 28-07-2018.
World Health Organization 2002 Cervical cancer screening in developing countries: Report of a WHO consultation. Program on Cancer Control. Last accessed on 28-03-2018.
Fahey MT, Irwiq L, Macaskill P: Meta-analysis of pap test accuracy. American Journal of Epidemiology 141(7):680–689, 1995
Bhattacharyya AK, Nath JD, Deka H: Comparative study between pap smear and Visual Inspection with Acetic acid (VIA) in screening of CIN and early cervical cancer. Journal of Mid-life Health 6(2):53–58, 2015
Sankaranarayanan R, Wesley R, Thara S, Dhakad N, Chandralekha B, Sebastian P, Chithrathara K, Parkin DM, Nair MK: Test characteristics of visual inspection with 4% acetic acid and Lugol’s Iodine in cervical cancer screening in Kerala. India. International Journal of Cancer 106(3):404–408, 2003
Belinson J, Pretorius R, Zhang W, Wu LY, Qiao YL, Elson P: Cervical cancer screening by simple visual inspection after acetic acid. Obstetrics & Gynecology 98(3):441–444, 2001
Chumworathayi B, Blumenthal PD, Limpaphayom KK, Kamsa-Ard S, Wongsena M, Supaatakorn P: Effect of single-visit VIA and cryotherapy cervical cancer prevention program in Roi Et, Thailand: a preliminary report. Journal of Obstetrics and Gynaecology Research 36(1):79–85, 2010
Sangwa-Lugoma G, Mahmud S, Nasr SH, Liaras J, Patrick KK, Tozin RR, Drouin P, Lorincz A, Ferenczy A, Franco EL: Visual inspection as a cervical cancer screening method in a primary health-care setting in Africa. International Journal of Cancer 119(6):1389–1395, 2006
Blumenthal P, Lauterbach M, Sellors J, Sankaranarayanan R: Training for cervical cancer prevention programs in low-resource settings: Focus on visual inspection with acetic acid and cryotherapy. International Journal of Gynecology & Obstetrics 89(2):S30–S37, 2005
Park SY, Follen M, Milbourne A, Rhodes H, Malpica A, MacKinnon N, MacAulay C, Markey MK, Richards-Kortum R: Automated image analysis of digital colposcopy for the detection of cervical neoplasia. Journal of Biomedical Optics 13(1):014029, 2008
Li W, Venkataraman S, Gustafsson U, Oyama JC, Ferris DG, Lieberman RW: Using acetowhite opacity index for detecting cervical intraepithelial neoplasia. Journal of Biomedical Optics 14(1):014020, 2009
Kim E, Huang X: A Data Driven Approach to Cervigram Image Analysis and Classification. In: Celebi M, Schaefer G Eds. Color Medical Image Analysis. Lecture Notes in Computational Vision and Biomechanics, Vol. 6. Dordrecht: Springer, 2013
T. Xu, E. Kim and X. Huang 2015 Adjustable AdaBoost classifier and pyramid features for image-based cervical cancer diagnosis. In Proc. IEEE 12th International Symposium on Biomedical Imaging, pages 281–285, New York, NY, USA
Kudva V, Prasad K, Guruvare S: Andriod Device-Based Cervical Cancer Screening for Resource-Poor Settings. Journal of Digital Imaging 31(5):646–654, 2018
Ji Q, Engel J, Craine E: Texture analysis for classification of cervix lesions. IEEE Transactions on Medical Imaging 19(11):1144–1149, 2000
Song D, Kim E, Huang X, Patruno J, Munoz-Avila H, Hein J: Multimodal entity coreference for cervical dysplasia diagnosis. IEEE Transactions on Medical Imaging 34(1):229–245, 2015
I. Claude, R. Winzenrieth, P. Pouletaut and J. C. Boulanger. Contour features for colposcopic images classification by articial neural networks. In Proc. 16th International Conference on Pattern Recognition, pages 771–774, Quebec City, Quebec, Canada, 2002.
V. V. Raad, Z. Xue and H. Lange. Lesion margin analysis for automated classification of cervicalcancer lesions. In Proc. SPIE Medical Imaging: Image Processing, volume 6144, pages 1–13, San Diego, California, United States, 2006.
Y. Jia, E. Shelhamer, J. Donahue, S. Karayev, J. Long, R. Girshick, S. Guadarrama and T. Darrell 2014 CAFFE: Convolutional Architecture for Fast Feature Embedding. arXiv:1408.5093. Last accessed on 28-03-2018.
A. Krizhevsky, I. Sutskever and G. E. Hinton. ImageNet classification with deep convolutional neural networks. In Proc. 25th International Conference on Neural Information Processing Systems, volume 1, page 1097–1105, Lake Tahoe, Nevada, 2012.
LeCun Y, Bottou L, Bengio Y, Haner P: Gradient-based learning applied to document recognition. Proceedings of the IEEE 86(11):2278–2324, 1998
P. Sermanet, D. Eigen, X. Zhang, M. Mathieu, R. Fergus and Y. LeCun. Over-Feat: Integrated recognition, localization and detection using convolutional networks. In Proc. International Conference on Learning Representations (ICLR2014), arXiv:1312.6229, 2014. Last accessed on 28-03-2018.
K. Simonyan and A. Zisserman 2014 Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556. Last accessed on 28-03-2018.
C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke and A. Rabinovich 2015 Going deeper with convolutions. In Proc. IEEE Conference on Computer Vision and Pattern Recognition, pages 1–9, Boston, MA, USA.
Zhanga W, Lia R, Dengb H, Wangc L, Lind W, Jia S, Shenc D: Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage 108:214–224, 2015
Gao Z, Wang L, Zhou L, Zhang J: HEp-2 cell image classification with deep convolutional neural networks. IEEE Journal of Biomedical and Health Informatics 21(2):416–428, 2017
Lequan Y, Hao C, Dou Q, Qin J, Heng PA: Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Transactions on Medical Imaging 36(4):994–1004, 2017
Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P: Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Computerized Medical Imaging and Graphics 61:2–13, 2017
Kudva V, Prasad K, Guruvare S: Automation of Detection of Cervical Cancer Using Convolutional Neural Networks. Critical Reviews in Biomedical Engineering 46(2):135–145, 2018
Ribeiro E, Andreas U, Wimmer G, Hafner M: Exploring deep learning and transfer learning for colonic polyp classification. Computational and Mathematical Methods in Medicine 2016:1–16, 2016
Abdolmanafi A, Duong L, Dahdah N, Cheriet F: Deep feature learning for automatic tissue classification of coronary artery using optical coherence tomography. Biomedical Optics Express 8(2):1203–1220, 2017
Abbas Q, Fondon I, Sarmiento A, Jiménez S, Alemany P: Automatic recognition of severity level for diagnosis of diabetic retinopathy using deep visual features. Medical & Biological Engineering & Computing 55(11):1959–1974, 2017
Beevi KS, Madhu SN, Bindu GR: Automatic mitosis detection in breast histopathology images using Convolutional Neural Network based deep transfer learning. Biocybernetics and Biomedical Engineering 39:214–223, 2019
Chen H, Ni D, Qin J, Li S, Yang X, Wang T, Heng PA: Standard plane localization in fetal ultrasound via domain transferred deep neural networks. IEEE Journal of Biomedical and Health Informatics 19(5):1627–1636, 2015
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J: Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Transactions on Medical Imaging 35(5):1299–1312, 2016
Paras L: Deep convolutional neural networks for endotracheal tube position and X-ray image classification: Challenges and opportunities. Journal of Digital Imaging 30(4):460–468, 2017
Phillip MC, Malhi HS: Transfer learning with convolutional neural networks for classification of abdominal ultrasound images. Journal of Digital Imaging 30(2):234–243, 2017
V. Vapnik 2013 The nature of statistical learning theory. Springer science & business media.
C. Zhang, S. Bengio and M. Hardt. Understanding deep learning requires rethinking generalization. arXiv:1611.03530v2 [cs.LG] 26 Feb 2017. Available online at https://arxiv.org/pdf/1611.03530.pdf. Last accessed on 15.06.2019
J. Wu, Q. Zhang and G. Xu. Tiny ImageNet challenge. Technical report, Stanford University, 2017. Available online at http://cs231n.stanford.edu/reports/2017/pdfs/930.pdf. Last accessed on 14-06-2019.
Acknowledgments
This publication is made possible by a sub-agreement from the Consortium for Affordable Medical Technologies (CAMTech) at Massachusetts General Hospital with funds provided by the generous support of the American people through the United States Agency for International Development (USAID grant number 224581). We would like to acknowledge the support of Mark Schiffman, M.D, M.P.H., Division of Cancer Epidemiology and Genetics, National Cancer Institute, USA, for providing us with cervix images. We would like to acknowledge the support of Dr. Suma Nair, Associate Professor, Community Medicine Department, Kasturba Medical College, Manipal, for facilitating the acquisition of images during the screening programs conducted.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclaimer
The contents are the responsibility of Manipal Academy of Higher Education and do not necessarily reflect the views of Massachusetts General Hospital, USAID or the United States Government.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kudva, V., Prasad, K. & Guruvare, S. Hybrid Transfer Learning for Classification of Uterine Cervix Images for Cervical Cancer Screening. J Digit Imaging 33, 619–631 (2020). https://doi.org/10.1007/s10278-019-00269-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-019-00269-1