Abstract
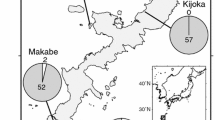
How phenotypic or genetic diversity is maintained in a natural habitat is a fundamental question in evolutionary biology. Flower color polymorphism in plants is a common polymorphism. Hepatica nobilis var. japonica on the Sea of Japan (SJ) side of the Japanese mainland exhibits within population flower color polymorphism (e.g., white, pink, and purple), while only white flowers are observed on the Pacific Ocean (PO) side. To determine the relationships between flower color polymorphism, within and among populations, and the genetic structure of H. nobilis var. japonica, we estimated the genetic variation using simple sequence repeat (SSR) markers. First, we examined whether cryptic lineages corresponding to distinct flower colors contribute to the flower color polymorphisms in H. nobilis var. japonica. In our field observations, no bias in color frequency was observed among populations on Sado Island, a region with high variation in flower color. Simple sequence repeat (SSR) analyses revealed that 18% of the genetic variance was explained by differences among populations, whereas no genetic variation was explained by flower color hue or intensity (0% for both components). These results indicate that the flower color polymorphism is likely not explained by cryptic lineages that have different flower colors. In contrast, populations in the SJ and PO regions were genetically distinguishable. As with the other plant species in these regions, refugial isolation and subsequent migration history may have caused the genetic structure as well as the spatially heterogeneous patterns of flower color polymorphisms in H. nobilis var. japonica.






Similar content being viewed by others
References
Arista M, Talavera M, Berjano R, Ortiz PL (2013) Abiotic factors may explain the geographical distribution of flower colour morphs and the maintenance of colour polymorphism in the scarlet pimpernel. J Ecol 101:1613–1622
Campbell DR, Waser NM, Melaendez-Ackerman EJ (1997) Analyzing pollinator-mediated selection in a plant hybrid zone: hummingbird visitation patterns on three spatial scales. Am Nat 149:295–315
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Cornuet J-M, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Crow JF (1986) Basic concepts in population, quantitative, and evolutionary genetics. W. H. Freeman and Company, New York
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from plant tissue. Focus 12:13–15
Edgar A (1936) Color variation in eastern north American flowers as exemplified by Hepatica actiloba. Rhodora 38:301–304
Frey FM (2004) Opposing natural selection herbivores and pathogens may maintain floral-color variation in Claytonia Virginica (Portulacaneae). Evolution 58:2426–2437
Frey FM (2007) Phenotypic integration and the potential for independent color evolution in a polymorphic spring ephemeral. Am J Bot 94:437–444
Fu DZ, Zhu GH (2001) Ranunculaceae L. In: Wu ZY, Raven P (eds) Flora of China, vol. 6. Science Press, Beijing and Missouri Botanical Garden Press, St. Louis, p 328
Futuyma DJ (2013) Evolution. Sinauer Associates, Sunderland
Gigord LDB, Rausher MD, Smithson A (2001) Negative frequency-dependent selection maintains a dramatic flower color polymorphism in the rewardless orchid Dactylorhiza sambucina (L.) Soò. PNAS 98:6253–6255
Goudet J (1995) FSTAT: A computer program to calculate F statics, version 1.2. J Hered 86:485–486
Horovitz A (1976) Edaphic factors and flower colour in the Anemoneae (Ranunculaceae). Plant Syst Evol 126:239–242
Jacobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jones KN, Reithel JS (2001) Pollinator-mediated selection on a flower color polymorphism in experimental populations of Antirrhinum (Scrophulariaceae). Am J Bot 88:447–454
Kalinowski ST (2005) HP-RARE 1 0: a computer program for performing arefaction on measures of allelic richness. Mol Ecol Notes 7:579–582
Kameoka S, Higashi H, Setoguchi H (2015) Development of polymorphic microsatellite loci in the perennial herb Hepatica nobilis var. japonica (Ranunculaceae). Appl Plant Sci. doi:10.3732/apps.1400114
Lande R (1975) The maintenance of genetic variability by mutation in a polygenic character with linked loci. Genet Res Camb 26:221–235
Luikart G, Allendorf FW, Cornuet JM, Sherwin WB (1998) Distortion of allele frequency distributions provides a test for recent population bottlenecks. Herdity 89:238–247
Maruyama T, Fuerst PA (1985) Population bottlenecks and nonequilibrium models in population genetics. 2. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111:675–689
Masel J, King OD, Maufhan H (2007) The loss of adaptive plasticity during long periods of environmental stasis. Am Nat 169:38–46
Matsumura S, Yokoyama J, Fukuda T, Maki M (2009) Origin of the disjunct distribution of flower colour polymorphism within Limonium wrightii (Plumbaginaceae) in the Ryukyu Archipelago. Biol J Linn Soc 97:709–717
Meirmans PG, Tienderen PHV (2004) Genotype and genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Moran NA (1992) The evolutionary maintenance of alternative phenotypes. Am Nat 139:971–989
Morgan MT, Schoen DJ (1997) Selection on reproductive characters: floral morphology in Asclepias syriaca. Heredity 79:433–441
Norton NA, Fernando MTR, Herlihy CR, Busch JW (2015) Reproductive character displacement shapes a spatially structured petal color polymorphism in Leavenworthia stylosa. Evolution 69:1191–1207
Ӧpik M, Moora M, Zobel M, Saks Ü, Wheatley R, Wright F, Daniell T (2008) High diversity of arbuscular mycorrhizal fungi in a boreal herb-rich coniferous forest. New Phytol 179:867–876
Peakall R, Smouse PE (2012) GenAlEx: genetic analysis in Excel. Population genetic software for teaching and research-an update, version 6.5. Bioinformatics 28:2537–2539
Piry S, Luikart G, Cornuet JM (1999) Bottleneck: a computer program for detecting recent reductions in the effective population size suing allele frequency data. Heredity 90:502–503
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, http://www.R-project.org/
Rausher MD (2008) Evolutionary transitions in floral color. Int J Plant Sci 169:7–21
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Schemske DW, Bierzychudek P (2001) Perspective: Evolution of flower color in the desert annual Lianthus parryae: Wright revisited. Evolution 55:1269–1282
Setoguchi H, Ohba H (1995) Phylogenetic relationships in Crossostylis inferred from restriction site variation of chloroplast DNA. J Plant Res 108:87–92
Subramaniam B, Rausher MD (2000) Balancing selection on a floral polymorphism. Evolution 54:691–695
Takahashi Y, Kawata M (2013) A comprehensive test for negative frequency-dependent selection. Pop Ecol 55:499–509
Takahashi Y, Takakura K, Kawata M (2015) Flower color polymorphism maintained by overdominant selection in Sisyrinchium sp. J Plant Res 128:933–939
Tani N, Maruyama K, Tomaru N, Uchida K, Araki M, Tsumura Y, Yoshimaru H, Ohba K (2003) Genetic diversity of nuclear and mitochondrial genomes in Pinus parviflora Sieb. & Zucc. (Pinaceae) populations. Herdity 91:510–518
Tsumura Y, Kado T, Takahashi T, Tani N, Ujino-Ihara T, Iwata H, Uchida K (2007) Genome scan to detect genetic structure and adaptive genes of natural populations of Cryptomeria japonica. Genetics 176:2393–2403
Volkova PA, Schanzer IA (2013) Colour polymorphism in common primrose (Primula vulgaris Huds.): many colours-many species? Plant Syst Evol 299:1075–1087
Weiss-Schneeweiss H, Schneeweiss GM, Stuessy TF, Mabuchi T, Park J-M, Jang C-G, Sun B-Y (2007) Chromosomal stasis in diploids contrasts with genome restricting in auto- and allopolyploid taxa of Hepatica (Ranunculaceae). New Phytol 174:669–682
Wessinger AC, Rausher MD (2012) Lessons from flower colour evolution on targets of selection. J Exp Bot 63:5741–5749
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kameoka, S., Sakio, H., Abe, H. et al. Genetic structure of Hepatica nobilis var. japonica, focusing on within population flower color polymorphism. J Plant Res 130, 263–271 (2017). https://doi.org/10.1007/s10265-016-0893-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-016-0893-1