Abstract
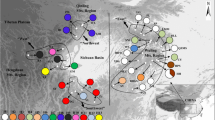
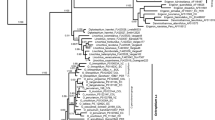
The dynamic changes in land configuration during the Quaternary that were accompanied by climatic oscillations have significantly influenced the current distribution and genetic structure of warm-temperate forests in East Asia. Although recent surveys have been conducted, the historical migration of forest species via land bridges and, especially, the origins of Korean populations remains conjectural. Here, we reveal the genetic structure of Lespedeza buergeri, a warm-temperate shrub that is disjunctively distributed around the East China Sea (ECS) at China, Korea, and Japan. Two non-coding regions (rpl32-trnL, psbA-trnH) of chloroplast DNA (cpDNA) and the internal transcribed spacer of nuclear ribosomal DNA (nrITS) were analyzed for 188 individuals from 16 populations, which covered almost all of its distribution. The nrITS data demonstrated a genetic structure that followed geographic boundaries. This examination utilized AMOVA, comparisons of genetic differentiation based on haplotype frequency/genetic mutations among haplotypes, and Mantel tests. However, the cpDNA data showed contrasting genetic pattern, implying that this difference was due to a slower mutation rate in cpDNA than in nrITS. These results indicated frequent migration by this species via an ECS land bridge during the early Pleistocene that then tapered gradually toward the late Pleistocene. A genetic isolation between western and eastern Japan coincided with broad consensus that was suggested by the presence of other warm-temperate plants in that country. For Korean populations, high genetic diversity indicated the existence of refugia during the Last Glacial Maximum on the Korean Peninsula. However, their closeness with western Japanese populations at the level of haplotype clade implied that gene flow from western Japanese refugia was possible until post-glacial processing occurred through the Korea/Tsushima Strait land bridge.





Similar content being viewed by others
References
Akiyama S (1988) A revision of the genus Lespedeza section Macrolespedeza (Leguminosae). Univ Mus Univ Tokyo Bull 33:1–170
Aoki K, Matsumura T, Hattori T, Murakami N (2006) Chloroplast DNA phylogeography of Photinia glabra (Rosaceae) in Japan. Am J Bot 93:1852–1858
Aoki K, Kato M, Murakami M (2009) Phylogeographical patterns of a generalist acorn weevil: insight into the biogeographical history of broadleaved deciduous and evergreen forests. BMC Evol Biol 9:103
Chang CS (2011) Illustrated encyclopedia of fauna and flora of Korea (in Korean). Woody plants. Ministry of Education, Science and Technology, vol 43. Designpost Press Co., Paju, p 260
Chung MY, López-Pujol J, Maki M, Moon MO, Hyun JO, Chung MG (2013) Genetic variation and structure within 3 endangered Calanthe species (Orchidaceae) from Korea: inference of population-establishment history and implications for conservation. J Hered 104:248–262
Chung MY, López-Pujol J, Chung MG (2014) Genetic homogeneity between Korean and Japanese populations of the broad-leaved evergreen tree Machilus thunbergii (Lauraceae): a massive post-glacial immigration through the Korea Strait or something else? Biochem Syst Ecol 53:20–28
Clark AG (1990) Inference of haplotypes from PCR-amplified samples of diploid populations. Mol Biol Evol 7:111–122
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene geologies. Mol Ecol 9:1657–1659
Comes HP, Abbott RJ (2001) Molecular phylogeography, reticulation, and lineage sorting in Mediterranean Senecio sect. Senecio (Asteraceae). Evolution 55:1943–1962
Corriveau JL, Coleman AW (1988) Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am J Bot 75:1443–1458
Crandall KA, Templeton AR (1993) Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 134:959–969
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772
Dobson M, Kawamura Y (1998) Origin of the Japanese land mammal fauna: allocation of extant species to historically-based categories. Quatern Res 37:385–395
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973
Excoffier L, Lischer H (2010) Arlequin Suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Han JE, Chung KH, Nemoto T, Choi BH (2010) Phylogenetic analysis of eastern Asian and eastern North American disjunct Lespedeza (Fabaceae) inferred from nuclear ribosomal ITS and plastid region sequences. Bot J Linn Soc 164:221–235
Hao QZ, Guo ZT, Qiao YS, Xu B, Oldfield F (2010) Geochemical evidence for the provenance of middle Pleistocene loess deposits in southern China. Quatern Sci Rev 29:23–24
Hare MP (2001) Prospects for nuclear gene phylogeography. Trends Ecol Evol 16:700–706
Harrison SP, Yu G, Takahara H, Prentice IC (2001) Palaeovegetation: diversity of temperate plants in East Asia. Nature 413:129–130
Hatusima S (1967) Lespedeza: sects. Macrolespedeza and Heterolespedeza from Japan, Corea and Formosa. Mem Fac Agric Kagoshima Univ 6:1–17
Hsu J, Kong ZC, Sun XJ, Tao JR, Du NQ (1967) A study of palaeobotany from Mount Jolmo Lunhma (in Chinese). In: Xizang Science Expedition Team, Chinese Academy (ed) Report of Scientific Expedition in the Mt. Jolmo Region (Quaternary Geology) 2. Science, Beijing, pp 93–103
Huang PH, Ohashi H, Nemoto T (2010) Lespedeza Michaux. In: Wu CY, Raven PH (eds) Flora of China, vol 10. Science Press, Beijing, pp 302–311
Keally CT (2005) Japanese Pleistocene landbridges and the earliest watercraft. Japanese Archaeology. http://www.t-net.ne.jp/~keally/MiddlePalaeol/landbridges.html. Accessed 12 Sept 2014
Kimura M (1996) Quaternary paleogeography of the Ryukyu Arc (in Japanese with English summary). J Geogr 105:259–285
Kimura M (2000) Paleography of the Ryukyu Island. Tropics 10:5–24
Lee TB (1965) The Lespedeza of Korea (1). Bull Seoul Natl Univ For 2:1–43
Lee TB (2003) Coloured flora of Korea. Hyangmunsa, Seoul, p 590
Lee JH, Lee DH, Choi BH (2013) Phylogeography and genetic diversity of East Asian Neolitsea sericea (Lauraceae) based on variation in chloroplast DNA sequences. J Plant Res 126:193–202
Lee JH, Lee DH, Choi IS, Choi BH (2014) Genetic diversity and historical migration patterns of an endemic evergreen oak, Quercus acuta, across Korea and Japan, inferred from nuclear microsatellites. Plant Syst Evol 300:1913–1923
Li EX, Yi S, Qiu YX, Guo JT, Comes HP, Fu CX (2008) Phylogeography of two East Asian species in Croomia (Stemonaceae) inferred from chloroplast DNA and ISSR fingerprinting variation. Mol Phylogenet Evol 49:702–714
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
Nakai T (1923) Notulæ ad Plantas Japoniæ et Coreæ XXX. Shokubutsugaku Zasshi 37:78
Nakai T (1927) Lespedeza of Japan and Korea. For Exp Stn Gov Gen Chosen 6:40
Nei M (1987) Molecular evolutionary genetics. Oxford University Press, New York
Okaura T, Quang ND, Ubakata M, Harada K (2007) Phylogeographic structure and late Quaternary population history of the Japanese oak Quercus mongolica var. crispula and related species revealed by chloroplast DNA variation. Genes Genet Syst 82:465–477
Ota H (1998) Geographic patterns of endemism and speciation in amphibians and reptiles of the Ryukyu Archipelago, Japan, with special reference to their paleogeographical implications. Res Popul Ecol 40:189–204
Ozaki K (1991) Late miocene and pliocene floras in Central Honshu, Japan. Bull Kanagawa Pref Mus Nat Sci Special issue:1–244
Pons O, Petit R (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Qi XS, Chen C, Comes HP, Sakaguchi S, Liu YH, Tanaka N, Sakio H, Qiu YX (2012) Molecular data and ecological niche modeling reveal a highly dynamic evolutionary history of the East Asian Tertiary relic Cercidiphyllum (Cercidiphyllaceae). New Phytol 196:617–630
Qi XS, Yuan N, Comes HP, Sakaguchi S, Qiu YX (2014) A strong ‘filter’ effect of the East China Sea land bridge for East Asia’s temperate plant species: inferences from molecular phylogeography and ecological niche modelling of Platycrater argua (Hydrangeaceae). BMC Evol Biol 14:41
Qiu YX, Sun Y, Zhang XP, Lee J, Fu CX, Comes HP (2009a) Molecular phylogeography of East Asian Kirengeshoma (Hydrangeaceae) in relation to quaternary climate change and landbridge configurations. New Phytol 183:480–495
Qiu YX, Qi XS, Tao XY, Fu CX, Naiki A, Comes HP (2009b) Population genetic structure, phylogeography, and demographic history of Platycrater arguta (Hydrangeaceae) endemic to East China and South Japan, inferred from chloroplast DNA sequence variation. Taxon 58:1226–1241
Qiu YX, Fu CX, Comes HP (2011) Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol Phylogenet Evol 59:225–244
Rambaut A, Drummond AJ (2007) Tracer. v. 1.5. http://beast.bio.ed.ac.uk/Tracer. Accessed 6 Feb 2014
Rieseberg LH, Soltis DE (1991) Phylogenetic consequences of cytoplasmic gene flow in plants. Evol Trends Plant 5:65–84
Rozas J, Rozas R (2001) DnaSP 3.53 DNA Sequence Polymorphism. University of Barcelona, Barcelona, Spain
Sakaguchi S, Qiu YX, Liu YH, Qi XS, Kim SH, Han JG, Takeuchi Y, Worth JRP, Yamasaki M, Sakurai S, Isagi Y (2012) Climate oscillation during the Quaternary associated with landscape heterogeneity promoted allopatric lineage divergence of a temperate tree Kalopanax septemlobus (Araliaceae) in East Asia. Mol Ecol 21:3823–3838
Sang T, Crawford DJ, Stuessy TF (1997) Chloroplast DNA phylogeny, reticulate evolution and biogeography of Paeonia (Paeoniaceae). Am J Bot 84:1120–1136
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot 94:275–288
Shi MM, Michalski SG, Welk E, Chen XY, Walter D (2014) Phylogeography of a widespread Asian subtropical tree: genetic east-west differentiation and climate envelope modelling suggest multiple glacial refugia. J Biogeogr 41:1710–1720
Sugahara K, Kaneko Y, Ito S, Yamanaka K, Sakio H, Hoshizaki K, Suzuki W, Yamanaka N, Setoguchi H (2011) Phylogeography of Japanese horse chestnut (Aesculus turbinata) in the Japanese Archipelago based on chloroplast DNA haplotypes. J Plant Res 124:75–83
Tao JR (1965) A late Eocene florula from the district Weinan of central Shensi (in Chinese with English summary). Acta Bot Sin 13:272–282
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wang J, Wu Y, Ren G, Guo Q, Liu J, Lascoux M (2011) Genetic differentiation and delimitation between ecologically diverged Populus euphratica and P. pruinosa. PLoS One 6:e26530
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, New York, pp 315–322
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058
Xu B, Gao XF, Zhang LB (2012) Analysis of DNA sequences of six plastid and nuclear genes suggests incongruence, introgression, and incomplete lineage sorting in the evolution of Lespedeza (Fabaceae). Mol Phylogenet Evol 62:346–358
Yokoyama J, Nakajima M, Nemoto T, Ohashi H (2000) Preliminary observations on flower visitors of Lespedeza subgenus Macrolespedeza in Korea. J Jpn Bot 75:248–256
Acknowledgments
The authors thank Prof. C. X. Fu of Zhejiang University for help in collecting plant materials. We are also grateful to our colleagues W. B. Cho, D. H. Lee, and I. S. Choi, at the Plant Systematics Laboratory of Inha University, for their discussions about statistical analysis and manuscript preparation. This research was supported by the National Research Foundation of Korea (NRF) (No. NRF–2015R1D1A1A01059886).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jin, DP., Lee, JH., Xu, B. et al. Phylogeography of East Asian Lespedeza buergeri (Fabaceae) based on chloroplast and nuclear ribosomal DNA sequence variations. J Plant Res 129, 793–805 (2016). https://doi.org/10.1007/s10265-016-0831-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-016-0831-2