Abstract
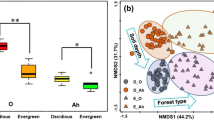
Soil microbes are considered to be a key determinant of the aboveground plant community. They are not distributed uniformly in the environment, and their activity, abundance, and ecosystem functioning could vary across localities, characterized by high β-diversity. Investigating factors that contribute to high β-diversity can help infer the possible mechanisms of microbial community assembly, and predict the scale and extent of impacts that soil microbes have on the plant community. Because soil systems consist of multiple horizons (i.e., vertical stratification) associated with different soil properties, complete understanding of high β-diversity requires consideration of both horizontal and vertical spatial structures of soil microbial communities. We studied the community composition of soil fungi from the O- and A-horizons in a Castanopsis-dominated temperate forest, and compared horizontal spatial autocorrelation in species composition between the two soil horizons (O- versus A-horizons). Pyrosequencing analysis yielded 67,129 sequencing reads, summed across all the 48 forest soil samples. Clustering analysis resulted in 597 molecular operational taxonomic units (OTUs), 68 % of which were identified as fungi, represented by four phyla. The Mantel test revealed that the O-horizon communities are spatially clustered, and the observed high β-diversity was driven not only by changes in OTUs present, but also by high turnover in identities of OTUs in soil samples. Furthermore, Mantel correlogram analysis showed that the O-horizon communities resembled each other in composition within the range of 50 m, whereas the A-horizon communities lacked such horizontal autocorrelation. These differences in the scale patchiness could arise from two processes: (1) that environmental conditions could show higher heterogeneity in finer scale at the A-horizon than at the O-horizon; and/or (2) dispersal could be more frequent at the O-horizon than the A-horizon. The present study suggests that either environmental filtering (i.e., the niche-based process) or dispersal limitation (i.e., neutral process) could characterize the observed patterns of spatial clustering in the soil fungal community.



Similar content being viewed by others
References
Abarenkov K, Nilsson RH, Larsson H-K, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vrålstad T, Liimatainen K, Peintner U, Kõljalg U (2010) The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol 186:281–285
Anderson IC, Cairney JWG (2004) Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ Microbiol 6:769–779
Auguspurger CK, Kelly CK (1984) Pathogen mortality of tropical tree seedlings: experimental studies of the effects of dispersal distance, seedling density, and light conditions. Oecologia 61:211–217
Bardgett RD (2005) The biology of soil. Oxford University Press, Oxford
Bissett A, Brown MV, Siciliano SD, Thrall P (2013) Microbial community responses to anthropogenically induced environmental change: towards a systems approach. Ecol Lett 16:128–139
Borcard D, Legendre P (2012) Is the Mantel correlogram powerful enough to be useful in ecological analysis? A simulation study. Ecology 93:1473–1481
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinform 10:421
Chase JM, Kraft NJB, Smith KG, Vellend M, Inouye BD (2011) Using null models to disentangle variation in community dissimilarity from variation in α-diversity. Ecosphere 2:2–11
Dickie IA, Xu B, Koide RT (2002) Vertical niche differentiation of ectomycorrhizal hyphae in soil as shown by T-RFLP analysis. New Phytol 156:527–535
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinform 27:2194–2200
Ettema CH, Wardle DA (2002) Spatial soil ecology. Trends Ecol Evol 17:177–183
Franklin RB, Mills AL (2003) Multi-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field. FEMS Microbiol Ecol 44:335–346
Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R (2008) Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5:235–237
Hibbett DS, Ohman A, Glotzer D, Nuhn M, Kirk P, Nilsson RH (2011) Progress in molecular and morphological taxon discovery in Fungi and options for formal classification of environmental sequences. Fungal Biol Rev 25:38–47
Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17:377–386
Izzo A, Agbowo J, Bruns TD (2005) Detection of plot-level changes in ectomycorrhizal communities across years in an old-growth mixed-conifer forest. New Phytol 166:619–630
Klironomos JN, Rillig MC, Allen MF (1999) Designing belowground field experiments with the help of semi-variance and power analyses. Appl Soil Ecol 12:227–238
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123
Legendre P, Legendre LFJ (1998) Numerical ecology. Elsevier Science, Amsterdam
Lilleskov EA, Bruns TD, Horton TR, Taylor DL, Grogan P (2004) Detection of forest stand-level spatial structure in ectomycorrhizal fungal communities. FEMS Microbiol Ecol 49:319–332
Lussenhop J, Wicklow DT (1984) Changes in spatial distribution of fungal propagules associated with invertebrate activity in soil. Soil Biol Biochem 16:601–604
Mangan SA, Schnitzer SA, Herre EA, Mack KML, Valencia MC, Sanchez EI, Bever JD (2010) Negative plant-soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466:752–755
Martiny JBH, Eisen JA, Penn K, Allison SD, Clair Horner-Devine M (2011) Drivers of bacterial β-diversity depend on spatial scale. Proc Nat Acad Sci USA 108:7850–7854
McGuire KL (2007) Common ectomycorrhizal networks may maintain monodominance in a tropical rain forest. Ecology 88:567–574
McGuire KL, Allison SD, Fierer N, Treseder KK (2013) Ectomycorrhizal-dominated boreal and tropical forests have distinct fungal communities, but analogous spatial patterns across soil horizons. PLoS ONE 8:e68278
Nara K (2006) Ectomycorrhizal networks and seedling establishment during early primary succession. New Phytol 169:169–178
Nilsson RH, Tedersoo L, Lindahl BD, Kjøller R, Carlsen T, Quince C, Abarenkov K, Pennanen T, Stenlid J, Bruns T, Larsson K-H, Kõljalg U, Kauserud H (2011) Towards standardization of the description and publication of next-generation sequencing datasets of fungal communities. New Phytol 191:314–318
O’Donnell AG, Young IM, Rushton SP, Shirley MD, Crawford JW (2007) Visualization, modelling and prediction in soil microbiology. Nature Rev Microbiol 5:689–699
Oksanen J, Blanchet GF, Kindt R, Legendre P, Minchin PB, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2012) Vegan: community ecology package. R package version 2.0–9. http://CRAN.R-project.org/package=vegan
Peay KG, Garbelotto M, Bruns TD (2010) Evidence of dispersal limitation in soil microorganisms: isolation reduces species richness on mycorrhizal tree islands. Ecology 91:3631–3640
Peay KG, Schubert MG, Nguyen NH, Bruns TD (2012) Measuring ectomycorrhizal fungal dispersal: macroecological patterns driven by microscopic propagules. Mol Ecol 21:4122–4136
R Development Core Team (2012) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Ranjard L, Dequiedt S, Chemidlin Prevost-Boure N, Thioulouse J, Saby NPA, Lelievre M, Maron PA, Morin FER, Bispo A, Jolivet C, Arrouays D, Lemanceau P (2013) Turnover of soil bacterial diversity driven by wide-scale environmental heterogeneity. Nature Comm 4:1434
Robinson CH, Szaro TM, Izzo AD, Anderson IC, Parkin PI, Bruns TD (2009) Spatial distribution of fungal communities in a coastal grassland soil. Soil Biol Biochem 41:414–416
Rosling A, Landeweert R, Lindahl BD, Larsson K-H, Kuyper TW, Taylor AFS, Finlay RD (2003) Vertical distribution of ectomycorrhizal fungal taxa in a podzol soil profile. New Phytol 159:775–783
Saetre P, Bååth E (2000) Spatial variation and patterns of soil microbial community structure in a mixed spruce–birch stand. Soil Biol Biochem 32:909–917
Sato H, Murakami N (2008) Reproductive isolation among cryptic species in the ectomycorrhizal genus Strobilomyces: population-level CAPS marker-based genetic analysis. Mol Phylo Evol 48:326–334
Sommer DD, Delcher AL, Salzberg SL, Pop M (2007) Minimus: a fast, lightweight genome assembler. BMC Bioinform 8:64
Tanabe AS (2012) Assams v0.1.2012.05.24. http://www.fifthdimension.jp/
Tanabe AS, Toju H (2013) Two new computational methods for universal DNA barcoding: a benchmark using barcode sequences of bacteria, archaea, animals, fungi, and land plants. PLoS ONE 8:e76910
Toju H, Tanabe AS, Yamamoto S, Sato H (2012) High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 7:e40863
Toju H, Yamamoto S, Sato H, Tanabe AS, Gilbert GS, Kadowaki K (2013) Community composition of root-associated fungi in a Quercus-dominated temperate forest: “co-dominance” of mycorrhizal and root-endophytic fungi. Ecol Evol 3:1281–1293
van der Heijden MGA, Bakker R, Verwaal J, Scheublin TR, Rutten M, van Logtestijn R, Staehelin C (2006) Symbiotic bacteria as a determinant of plant community structure and plant productivity in dune grassland. FEMS Micro Ecol 56:178–187
van der Heijden MGA, Bardgett R, van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11:296–310
Vilgalys R, Gonzalez D (1990) Ribosomal DNA restriction fragment length polymorphisms in Rhizoctonia solani. Phytopathol 80:151–158
Walker TS, Baisi HP, Grotewold E, Vivanco JM (2003) Root exudation and rhizosphere biology. Plant Physiol 132:44–51
Weber CF, Vilgalys R, Kuske CR (2013) Changes in fungal community composition in response to elevated atmospheric CO2 and nitrogen fertilization varies with soil horizon. Front Microbiol 4:78
Acknowledgments
We thank T Miki and two anonymous reviewers for comments that improved the manuscript. Financial support was provided through the Funding Program for Next Generation World-Leading Researchers of Cabinet Office, the Japanese Government (GS014) to HT.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kadowaki, K., Sato, H., Yamamoto, S. et al. Detection of the horizontal spatial structure of soil fungal communities in a natural forest. Popul Ecol 56, 301–310 (2014). https://doi.org/10.1007/s10144-013-0424-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10144-013-0424-z