Abstract
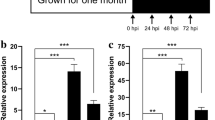
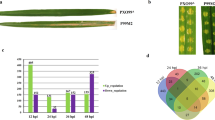
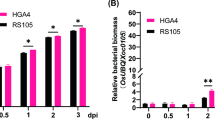
Bacterial leaf streak (BLS) is now the fourth-most devastating disease in rice. Dular and H359 are two indica rice varieties with contrasting responses to BLS. Dular displays high resistance, while H359 is susceptible. In this study, RNA-seq was used to examine the early molecular processes deployed during the resistance response of Dular and H359 at different times after inoculation. Differentially expressed gene (DEG) analysis identified 3031 genes in Dular and 7161 in H359 that were modulated in response to infection after 12 and 24 h. There were significantly more DEGs in H359 than in Dular, and there were significantly more downregulated genes than upregulated genes. Gene ontology (GO) and KEGG enrichment analyses revealed a similar set of GO terms and KEGG pathways enriched in both varieties. However, KEGG analysis of upregulated DEGs revealed that some phenylpropane metabolism-related pathways were specially enriched in Dular. Further comparison and analysis showed that the numbers of resistance-related DEGs in the two varieties were significantly reduced at 24 h compared with 12 h after BLS infection and genes critically involved in conferring resistance during the early stage mainly included WRKY transcription factors, receptor kinases and disease, exocyst, MAPK signalling pathway and hormones related genes. Our study suggests that resistance-related genes may play an important role at an early stage of infection and phenylpropane metabolism related genes may partly response for BLS resistance of Dular, thus providing valuable information for future studies on the molecular mechanisms of BLS resistance in rice.





Similar content being viewed by others
References
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Camargo-Ramírez R, Val-Torregrosa B, Segundo BS (2018) MiR858-mediated regulation of flavonoid-specific MYB transcription factor genes controls resistance to pathogen infection in Arabidopsis. Plant Cell Physiol 59(1):190–204
Chen CH, Cheng W, Huang XM, Zhang DP, Lin XH (2006) Major QTL conferring resistance to rice bacterial leaf streak. Agric Sci China 5:216–220
Chen XJ, Chen Y, Zhang LN, Xu B, Zhang JH, Chen ZX, Tong TH, Zuo SM, Xu JY (2016) Overexpression of OsPGIP1 enhances rice resistance to sheath blight. Plant Dis 100:388–395
Cheng XJ, He B, Chen L, Xiao SQ, Fu J, Chen Y, Yu TQ, Cheng ZQ, Feng H (2016) Transcriptome analysis confers a complex disease resistance network in wild rice oryza meyeriana against Xanthomonas Oryzae pv. Oryzae. Sci Rep 6(1):38215
Dixon RA, Achnine L, Kota P, Liu CJ, Reddy MSS, Wang L (2002) The phenylpropanoid pathway and plant defence—a genomics perspective. Mol Plant Pathol 3(5):371–390
Du H, Wu N, Fu J, Wang SP, Li XH, Xiao JH, Xiong LZ (2012) a GH3 family member OsGH3-2 modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J Exp Bot 63:6467–6480
Feng CS, Zhang X, Wu T, Yuan B, Ding XH, Yao FY, Chu ZH (2016) The polygalacturonase-inhibiting protein 4 (OsPGIP4) a potential component of the qblsr5a locus confers resistance to bacterial leaf streak in rice. Planta 243:1297–1308
Formentin E, Sudiro C, Perin G, Riccadonna S, Barizza E, Baldoni E, Lavezzo E, Stevanato P, Sacchi GA, Fontana P, Toppo S, Morosinotto T, Zottini M, Lo Schiavo F (2018) Transcriptome and cell physiological analyses in different rice cultivars provide new insights into adaptive and salinity stress responses. Front Plant Sci 9:204
Fu J, Liu HB, Li Y, Yu HB, Li XH, Xiao JH, Wang SH (2011) Manipulating broad-spectrum disease resistance by suppressing pathogen-induced Auxin accumulation in rice. Plant Physiol 155:589–602
Gimenez-lbanez S, Solana R (2013) Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front Plant Sci 4:72
Hutin M, Césari S, Chalvon V, Michel C, Tran TT, Boch J, Koehnik R, Szurek B, Kroj T (2016) Ectopic activation of the rice NLR heteropair RGA4/RGA5 confers resistance to bacterial blight and bacterial leaf streak diseases. Plant J 88:43–55
Ishihara A, Hashimoto Y, Tanaka C, Dubouzet JG, Nakao T, Matsuda F, Nishioka T, Miyagawa H, Wakasa K (2008) The tryptophan pathway is involved in the defense responses of rice against pathogenic infection via serotonin production. Plant J 54(3):481–495
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Kalmbach L, Hématy K, Bellis DD, Barberon M, Fujita S, Ursache R, Daraspe J, Geldner N (2017) Transient cell-specific EXO70A1 activity in the CASP domain and casparian strip localization. Nat Plants 3:17058
Kim D, Langmead B, Salzberg SL (2015) HISTA: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Kulich I, Vojtíková Z, Sabol P, Ortmannová J, Nedela V, Tihlarikova E, Žárský V (2018) Exocyst subunit EXO70H4 has a specific role in callose synthase secretion and silica accumulation. Plant Physiol 176:2040–2051
Li JL, Pan YH, Guo HF, Zhou L, Yang S, Zhang ZY, Yang JZ, Zhang HL, Li GG, Zeng YW, Li ZC (2018) Fine mapping of QTL qCTB10-2 that confers cold tolerance at the booting stage in rice. Theor Appl Genet 131(1):157–166
Li B, Ferreira MA, Huang M, Camargos LF, Yu X, Teixeira RM, Carpinetti PA, Mendes GC, Gouveia-Mageste BC, Liu C, Pontes CSL, Brustolini OJB, Martins LGC, Melo BP, Duarte CEM, Shan L, He P, Fontes EPB (2019a) The receptor-like kinase NIK1 targets FLS2/BAK1 immune complex and inversely modulates antiviral and antibacterial immunity. Nat Commun 10:4996
Li YG, Yang YH, Hu YL, Liu HL, He M, Yang ZY, Kong FJ, Liu X, Hou XL (2019b) Della and EDS1 form a feedback regulatory module to fine-tune plant growth-defense tradeoff in Arabidopsis. Mol Plant 12:1485–1498
Lin CY, Trinh NN, Fu SF, Hsiung YC, Chia LC, Lin CW, Huang HJ (2013) Comparison of early transcriptome responses to copper and cadmium in rice roots. Plant Mol Biol 81:507–522
Liu N, Hake K, Wang W, Zhao T, Romeis T, Tang DZ (2016) Calcium-dependent protein kinase5 associates with the truncated NLR protein TIR-NBS2 to contribute to EXO70B1-mediated immunity. Plant Cell 29:746–759
Liu QN, Ning YS, Zhang YX, Yu N, Zhao CD, Zhan XD, Wu WX, Chen DB, Wei XJ, Wang GL, Cheng SH, Cao LY (2017) OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell 29:345–359
Mao H, Nakamura M, Viotti C, Grebe M (2016) A framework for lateral membrane trafficking and polar tethering of the PEN3 ATP-binding cassette transporter. Plant Physiol 172:2245–2260
Park CJ, Ronald PC (2012) Cleavage and nuclear localization of the rice XA21 immune receptor. Nat Commun 3:920
Peng Y, Bartley LE, Chen XW, Dardick C, Chern M, Ruan R, Canlas PE, Ronald PC (2008) OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv oryzae in rice. Mol Plant 1:446–458
Ren DY, Rao YC, Huang LC, Leng YJ, Hu J, Lu M, Zhang GH, Zhu L, Gao ZY, Dong GJ, Guo BL, Qian Q, Zeng DL (2016) Fine mapping identifies a new qtl for brown rice rate in rice (oryza sativa l.). Rice 9(1):1–10
Ryu HS, Han M, Lee SK, Cho J, Ryoo N, Heu S, Lee YH, Bhoo SH, Wang GL, Hahn TR, Jeon JS (2006) A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep 25:836–847
Seo YS, Sriariyanun M, Wang L, Pfeiff J, Phetsom J, Lin Y, Jung KH, Chou HH, Bogdanove A, Ronald P (2008) A two-genome microarray for the rice pathogens Xanthomonas oryzae pv oryzae and X oryzae pv oryzicola and its use in the discovery of a difference in their regulation of hrp genes. BMC Microbiol 8:99
Seo YU, Chern MS, Bartley LE, Han M, Jung KH, Lee I, Walia H, Richter T, Xu X, Cao P, Bai W, Ramanan R, Amonpant F, Arul L, Canlas PE, Ruan R, Park CJ, Chen XW, Hwang S, Jeon JS, Ronald PC (2011) Towards establishment of a rice stress response interactome. PLoS Genet 7:e1002020
Stegmann M, Anderson RG, Ichimura K, Pecenkova T, Reuter P, Zarsky V, McDowell JM, Shirasu K, Trujillo M (2012) The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24:4703–4716
Sun LJ, Zhang HJ, Li DY, Huang L, Hong YB, Ding SH, Nelson RS, Zhou XP, Song FM (2013) Functions of rice NAC transcriptional factors ONAC122 and ONAC131 in defense responses against Magnaporthe grisea. Plant Mol Biol 81:41–56
Takatsuji H, Hayashi N (2017) Broad-spectrum blast resistance: harnessing a natural allele of a transcription factor in rice. Mol Plant 10:1144–1146
Tang DZ, Wu WR, Li W, Lu H, Worland AJ (2000) Mapping of QTLS conferring resistance to bacterial leaf streak in rice. Theor Appl Genet 101:286–291
Tian L, Shi SH, Nasir F, Chang CL, Li WQ, Tran LSP (2018) Comparative analysis of the root transcriptomes of cultivated and wild rice varieties in response to Magnaporthe oryzae infection revealed both common and species-specific pathogen responses. Rice 11:26
Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M (2007) Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc Natl Acad Sci U S A 104:1075–1080
Wamaitha MJ, Yamamoto R, Wong HL, Kawasaki T, Kawano Y, Shimamoto K (2012) OsRap26 transcription factor contributes to rice innate immunity through its interaction with receptor for activated kinase-C 1 (RACK1). Rice 5:35
Wang R, Lu LX, Pan XB, Hu ZL, Ling F, Yan Y, Liu YM, Lin YJ (2015) Functional analysis of OsPGIP1 in rice sheath blight resistance. Plant Mol Biol 87:181–191
Wang W, Feng BM, Zhou JM, Tang DZ (2019a) Plant immune signaling: advancing on two frontiers. J Integr Plant Biol 10:1111
Wang R, Wang GL, Ning Y (2019b) PALs: emerging key players in broad-spectrum disease resistance. Trends Plant Sci 24(9):78–787
Weston LA, Mathesius U (2013) Flavonoids: their structure, biosynthesis and role in the Rhizosphere, including Allelopathy. J Chem Ecol 39(2):283–297
Xiao CL, Xie SQ, Xie QB, Liu ZY, Xing JF, Ji KK, Tao J, Dai LY, Luo F (2018) N6-Methyladenine DNA modification in Xanthomonas oryzae pv oryzicola genome. Sci Rep 8:6272
Xie C, Mao XZ, Huang JJ, Ding Y, Wu JM, Dong S, Kong L, Gao G, Li CY, Wei LP (2011) KOBAS 20: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:316–322
Xie XF, Chen ZW, Cao JL, Guan HZ, Lin DG, Li CL, Lan T, Duan YL, Mao DM, Wu WR (2014) Toward the positional cloning of qBlsr5a a QTL underlying resistance to bacterial leaf streak using overlapping sub-CSSLs in rice. PLoS One 9:e95751
Xu XH, Wang C, Li SX, Su ZZ, Zhou HN, Mao LJ, Feng XX, Liu PP, Chen X, Snyder JH, Kubicek CP, Zhang CL (2015) Lin FC friend or foe: differential responses of rice to invasion by mutualistic or pathogenic fungi revealed by RNA-seq and metabolite profiling. Sci Rep 5:13624
Yadav V, Wang ZY, Wei CH, Amo A, Ahmed B, Yang XZ, Zhang X (2020) Phenylpropanoid pathway engineering: An emerging approach towards plant defense. Pathogens 9(4):E312
Yamada S, Kano A, Tamaoki D, Miyamoto A, Shishido H, Miyoshi S, Taniguchi S, Akimitsu K, Gomi K (2012) Involvement of OsJAZ8 in jasmonate-induced resistance to bacterial blight in rice. Plant Cell Physiol 53:2060–2072
Yamanaka N, Watanabe S, Toda K, Hayashi M, Fuchigami H, Takahashi R, Harada K (2005) Fine mapping of the ft1 locus for soybean flowering time using a residual heterozygous line derived from a recombinant inbred line. Theor Appl Genet 110(4):634–639
Yang W, Zhang BG, Qi GH, Shang LY, Liu HF, Ding XH, Chu ZH (2019) Identification of the Phytosulfokine receptor 1 (OsPSKR1) confers resistance to bacterial leaf streak in rice. Planta 250:1603–1612
Ye H, Du H, Tang N, Li XH, Xiong LZ (2009) Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol Biol 71:291–305
Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, Yamane H, Shimono M, Sugano S, Takatsuji H, Kaku H, Minami E, Nishizawa Y (2013) WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Exp Bot 64:5085–5097
Yoo YH, Nalini Chandran AK, Park JC, Gho YS, Lee SW, An G, Jung KH (2017) OsPhyb-mediating novel regulatory pathway for drought tolerance in rice root identified by a global RNA-seq transcriptome analysis of rice genes in response to water deficiencies. Front Plant Sci 8:580
Yuan M, Ke YG, Huang RY, Ma L, Yang ZY, Chu ZH, Xiao JH, Li XH, Wang SP (2016) A host basal transcription factor is a key component for infection of rice by TALE-carrying bacteria. Elife 5:e19605
Zhai K, Deng YW, Liang D, Tang J, Liu J, Yan BX, Yin X, Lin H, Chen FD, Yang DY, Xie Z, Liu JY, Li Q, Zhang L, He ZH (2019) RRM transcription factors interact with NLRs and regulate broad-spectrum blast resistance in rice. Mol Cell 74:996–1009
Zhao T, Rui L, Li J, Nishimura MT, Vogel JP, Liu N, Liu S, Zhao YZ, Dangl JF, Tang DZ (2015) A truncated NLR protein TIR-NBS2 is required for activated defense responses in the EXO70B1 mutant. PLoS Genet 11:e1004945
Zhao HJ, Wang XY, Jia YL, Minkenberg B, Wheatley M, Fan JB, Jia MH, Famoso A, Edwards JD, Wamishe Y, Valent B, Wang GL, Yang YN (2018) The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat Commun 9:2039
Zhao YF, Wu GH, Shi H, Tang DZ (2019) Receptor-like kinase 902 associates with and phosphorylates Brassinosteroid-signaling kinase 1 to regulate plant immunity. Mol Plant 12:57–70
Funding
This work was supported in part by the National Key R&D Programme of China (2017YFD0100100), Fujian Provincial Natural Science Foundation, China (2017 J01438; 2019 J01424), Youth Technology Innovation Team of the Fujian Academy of Agricultural Sciences (No. STIT2017-3-3), Regional Development Project of Fujian, China (2018N3011), and General Project of the Fujian Academy of Agricultural Sciences (No. A2017-13).
Author information
Authors and Affiliations
Contributions
Zhiwei Chen, Shengping Li and Dingzhong Tang conceived and designed the research. Lin Lu and Dewei Yang carried out most experiments. Lin Lu, Shengping Li and Dewei Yang wrote the manuscript. Lin Lu, Shengping Li and Dewei Yang analysed the data. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict interest
These authors declare that they have no conflict interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lu, L., Yang, D., Tang, D. et al. Transcriptome analysis of different rice cultivars provides novel insights into the rice response to bacterial leaf streak infection. Funct Integr Genomics 20, 681–693 (2020). https://doi.org/10.1007/s10142-020-00744-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-020-00744-x