Abstract
Background
ADAMTS5 has been reported to be involved in the progression of several human tumors. Nevertheless, the role of ADAMTS5 in gastric cancer (GC) remains poorly defined.
Methods
ADAMTS5 expression levels were analyzed by quantitative real-time PCR (qRT-PCR) and immunohistochemistry (IHC) in GC cell lines and tissues, and the correlations between ADAMTS5 expression and clinicopathological features and survival were also examined. In vitro assays, including transwell assays, wound healing assays and cell adhesion assays, were employed to further explore the biological functions of ADAMTS5. A MAP kinase pathway microarray was used to identify the underlying mechanisms. The expression of ADAMTS5 and ETS1 and the microvessel density (MVD) were also analyzed using IHC to determine correlations with angiogenesis in GC.
Results
ADAMTS5 expression was downregulated in gastric cancer tissues. Low expression of ADAMTS5 was associated with gender, histological type, degree of differentiation, M stage, TNM stage and vascular invasion, and was also an independent indicator of a poor prognosis for patients with GC. ADAMTS5 overexpression markedly inhibited GC cell migration and invasion and enhanced cell adhesion to the extracellular matrix (ECM), whereas knockdown of ADAMTS5 exerted the opposite effects. Furthermore, the ADAMTS5 expression status was negatively correlated with ETS1 expression and MVD.
Conclusion
ADAMTS5 is downregulated in GC and suppresses tumor metastasis and angiogenesis by inhibiting ETS1-mediated changes in MVD and potentially acts as a novel prognostic marker and a potential therapeutic target in human GC.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is the most common upper gastrointestinal tumor and the third leading cause of tumor-related deaths worldwide. An estimated 951,600 new gastric cancer cases and 723,100 related deaths occurred in 2012 [1]. Although advances in treatment have been achieved, patients with GC still experience a low survival rate due to the high rates of local and distant metastasis and relapse. Hence, we aimed to explore the molecular mechanism underlying metastasis by identifying biomarkers that might facilitate the development of precise treatment strategies for GC.
The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family includes 19 secreted zinc metalloproteases that have been found to be involved in various biological and pathological processes, including tissue morphogenesis, inflammation, angiogenesis and cancer [2]. To date, 10 of the 19 members have been confirmed to be involved in cancer as a tumor suppressor or oncogene [3, 4]. In different microenvironments, the same molecule exerts a distinct function on tumorigenesis, such as ADAMTS1 [5,6,7]. ADAMTS5 was identified as a key enzyme in osteoarthritis by degrading aggrecan as an aggrecanase after its discovery in 1999 [8]. Based on recent accumulating evidence, ADAMTS5 also plays important roles in normal physiology as well as pathologies such as cancer [9]. Downregulation or hypermethylation of ADAMTS5 inhibited its tumor suppressor function in mouse melanoma [10], human hepatocellular carcinoma (HCC) [11] and human colorectal cancer [12]. Meanwhile, overexpression of ADAMTS5 has also been reported in human glioblastomas and stimulates cell invasion [13]. Thus, ADAMTS5 probably has different roles or is involved in different molecular mechanisms in various tumors. However, the exact role of ADAMTS5 in human GC remains unknown.
In the present study, the downregulation of ADAMTS5 was correlated with clinicopathological features and functioned as an independent indicator of a poor prognosis for patients with GC. Furthermore, a variety of in vitro assays revealed that ADAMTS5 significantly suppressed tumor invasion and migration, potentially by reducing ETS1-mediated changes in the MVD.
Materials and methods
Patients and tissue samples
Formalin-fixed and paraffin-embedded tissues were collected from 176 patients with gastric carcinoma who underwent surgical resection at The Sixth Affiliated Hospital of Sun Yat-Sen University, Guangzhou, China, from December 2007 to March 2012. The samples were used to construct a tissue microarray and for immunohistochemistry (IHC). Patients were followed until death or to December 31, 2017, except for patients who were lost to follow-up. Overall survival (OS) was defined as the interval between the date of surgery and the date of death or last follow-up visit. GC staging was classified according to the 7th Edition of the AJCC Gastric Cancer Staging System. An independent set of 30 pairs of fresh-frozen gastric cancer and paired noncancerous gastric tissues preserved during the same time span were used for mRNA extraction. Informed consent was obtained from all patients, and the study was approved by the Research Ethics Committee of Sun Yat-Sen University and complied with the principles of the Declaration of Helsinki.
Immunohistochemistry
ADAMTS5, CD31 and ETS1 expression were detected by IHC staining using a Biotin-Streptavidin HRP Detection System (ZSGB-Bio, China), as previously described [14]. Briefly, sections were incubated with primary rabbit antibodies against ADAMTS5 (ab41037, Abcam; 1:200), CD31 (77699S, CST; 1:100) and ETS1 (ab26096, Abcam; 1:100) overnight at 4 °C. The primary antibody diluent was used as a negative control. Specimens were developed with DAB and counterstained with hematoxylin. Sections were photographed under a microscope and analyzed by two pathologists who were blinded to the identity of the pathological materials. ADAMTS5 and ETS1 expression were scored using a semi-quantitative system, the staining index (SI), as previously described [14]. Receiver operating characteristic (ROC) curves were analyzed to select cutoff scores (3.5 for ADAMTS5, 4.5 for ETS1). For the quantification of MVD, the number of blood vessels exhibiting positive staining for CD31 was recorded in each complete spot of the microarray, and the cutoff score was 62.5.
Cell lines and cell culture
Human GC cell lines AGS, MKN45, SGC7901, BGC823, MGC803, MKN1, SNU1 and HGC27, and human normal gastric mucosal cells GES1 and HEK 293T cells were obtained from the Type Culture Collection Cell Bank of the Chinese Academy of Sciences Committee (Shanghai, China). AGS and HEK 293T cells were cultured in DMEM/F12 and DMEM, respectively. The other cell lines were cultured in RPMI 1640 medium. All media were supplemented with 10% fetal bovine serum (FBS). Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2. For the demethylation analysis, AGS and MKN45 cell lines expressing methylated ADAMTS5 were treated with 2 µM 5-aza-2′-deoxycytidine (5-Aza-dC, Sigma, USA) for 3 days. The cells in the 5-aza-dC/trichostatin A(TSA) group were treated with 2 µM 5-aza-dC for 3 days followed by 2 µM TSA for 24 h. Cells treated with an equivalent concentration of dimethylsulfoxide (DMSO) were used as controls.
Bisulfite genomic sequencing and methylation-specific PCR
Genomic DNA was extracted from cell lines and tissues using a gDNA purification kit (Promega, USA) and then modified with bisulfate using a DNA methylation golden kit (ZYMO, USA), according to the manufacturers’ protocols. Bisulfite-modified genomic DNA was amplified by polymerase chain reaction (PCR) using bisulfite genomic sequencing (BGS) and methylation-specific PCR (MSP) primers. The primers for BGS were 5-AATTGTTATTGTAGGATGGTATGT-3 (forward) and 5-TTTCCTTAACTCTACTAAAACCAA-3 (reverse). The primers for MSP were methylated, 5-TTCGTATTTTTATAGCGTTCGC-3 (forward), 5-GTATCTATATCCCTTCGACTACGA-3 (reverse), and unmethylated, 5-TTTGTATTTTTATAGTGTTTGTGT-3 (forward), 5-ATATCTATATCCCTTCAACTACAA-3 (reverse).
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cell lines and tissues using TRIzol reagent (Invitrogen, USA), according to the manufacturer’s instructions. Reverse transcription and qRT-PCR were performed as previously described [14]. β-Actin was used as an endogenous control. The primer sequences used in the qRT-PCR analysis were ADAMTS5, 5-GGCCTCCATCGCCAATAGG-3 (forward), 5-GGATAGCTGCATCGTAGTGCT-3 (reverse); ETS1, 5-GATAGTTGTGATCGCCTCACC-3 (forward), 5-GTCCTCTGAGTCGAAGCTGTC-3 (reverse); and β-actin, 5-ACAATGTGGCCGAGGACTTT-3 (forward), 5-GGAGAGGACTGGGCCATTCT-3 (reverse).
Western blot assay
Total proteins were extracted from cell lysates using T-PER Tissue Protein Extraction Reagent (Thermo Fisher, USA) supplemented with protease and phosphatase inhibitors (Roche, Switzerland). Protein concentrations were measured using a BCA protein assay kit (Beyotime, China). Proteins were separated by 10% SDS-PAGE and then transferred to PVDF membranes (Millipore, USA). Membranes were then blocked with 5% skim milk at room temperature for 1 h and incubated with the primary antibodies overnight at 4 °C, followed by an incubation with corresponding IRDye-conjugated secondary antibodies (LI-COR Biosciences, USA) for 1 h at room temperature. Bands were detected using the Odyssey Infrared Imaging System (LI-COR Biosciences, America). Cells were plated in culture dishes containing 5 mL of growth medium and incubated until they were approximately 90% confluent to analyze the levels of the secreted ADAMTS5 protein in the culture medium. The growth medium was discarded, and cells were washed with phosphate-buffered saline. Afterwards, a serum-reduced medium (5 mL) supplemented with 0.2% fetal bovine serum was added, and cells were continually incubated in a 5% CO2 atmosphere at 37 °C for 24 h. The conditioned media were harvested and centrifuged at 3000 rpm for 5 min. The supernatants of centrifuged conditioned media were collected and freeze-dried using a vacuum freeze dryer. The precipitate was mixed with T-PER Tissue Protein Extraction Reagent, and the protein concentration was quantified using a protein assay reagent (BCA, Beyotime, China), followed by western blot analysis. Protein expression levels were normalized to β-actin levels in the cell lysates.
Establishment of cell lines displaying stable ADAMTS5 overexpression or knockdown
The complete open reading frame (ORF) of ADAMTS5 (NM_007038.4) was amplified by PCR using the following primers: 5-TCCGACACTTGACTCAATCC-3 (forward) and 5-GCATGACTTTCTGTGCGTTAG-3 (reverse). The PCR product was purified and then ligated into a linearized pCDH-CMV-MCS-EF1-copGFP plasmid (SBI, USA). The recombinant plasmid was transformed into competent Escherichia coli DH5α cells (Takara, Japan) for amplification. The identity of the plasmid was confirmed by sequencing. HEK 293T cells were co-transfected with the empty lentiviral plasmid or the ADAMTS5-overexpressing lentiviral plasmid with the packing plasmid and envelope plasmid to produce lentiviruses, according to the manufacturer’s instructions. For infection, a lentiviral suspension containing approximately 4 × 106 lentiviral particles (MOI = 10) and polybrene (5 µg/mL) were added to AGS or MKN45 cells. The infection efficiency was confirmed to exceed 90% for further experiments, as determined by the percentage of GFP-positive cells under a microscope.
For ADAMTS5 knockdown, an ADAMTS5-specific shRNA lentiviral vector and control shRNA vector were constructed using the GV248 vector (Genechem Co. Ltd., China). The shRNA sequence targeting ADAMTS5 was 5-AGACTAGATTCACTGCCTA-3, and the control shRNA sequence was 5-TTCTCCGAACGTGTCACGT-3. Lentiviral packaging and infection were performed as mentioned above using SGC7901 and BGC823 cells.
Transwell migration and invasion assays
The migration and invasion assays were performed using a transwell chamber with 8-µm filter inserts (BD Biosciences, USA) as previously described [14]. Briefly, for the migration assay, 5 × 104 cells were resuspended in 120 µL of serum-free medium and plated in the upper chamber and 600 µL of medium containing 10% FBS were added to the lower chamber. After a 24-h incubation at 37 °C, cells in the upper chamber were gently removed, while cells that traversed to the lower membrane surface were fixed with 4% paraformaldehyde and stained with 4′,6-diamidino-2-phenylindole (DAPI, 100 ng/mL). Cells were imaged and counted in five random 200× fields under a fluorescence microscope (Olympus, Japan). For the invasion assay, 5 × 104 cells resuspended in 120 µL of serum-free medium were added to the Matrigel-coated membrane (BD Biosciences, USA) in the upper chamber. Invasion was determined after a 36-h incubation. Each assay was performed in triplicate.
Wound healing assay
A wound healing assay was employed to further investigate cancer cell migration. First, 5 × 105 cells were seeded in 6-well plates and the cell monolayer was scratched in a straight line with a sterile 100-µL pipette tip at 90% confluence. After a 36-h incubation with medium containing 1% FBS, the wounds were observed under a microscope and photographed at 100× magnification at 0, 24, and 36 h. The wound healing areas were measured using ImageJ software (National Institutes of Health, USA) and calculated as the percentage of wound closure, with the initial wound width defined as 100%. All experiments were repeated three times in triplicate.
Cell adhesion assay
After 96-well plates coated with 50 µL of 20 µg/mL fibronectin (BD Biosciences, USA) were incubated at 4 °C overnight, the plates were washed with PBS twice and then blocked with 100 µL of 1% BSA for 1 h. Then, 2 × 104 cells were added to the coated wells and incubated at 37 °C for 45 min. After the non-adherent cells were removed by two washes with PBS, the adherent cells were then measured using two independent methods. The adherent cells were fixed with 4% paraformaldehyde and stained with DAPI (100 ng/mL). Cells were imaged and counted in five random 200× fields under a fluorescence microscope (Olympus, Japan). Meanwhile, the initial input cells and the adherent cells were measured using the MTS assay. Each well was treated with 20 µL of MTS reagent (Promega, USA), and the plates were incubated at 37 °C for 2 h. The absorbance of each well was measured at 490 nm. The results were expressed as the percentage of adherent cells, with initial input cells set to 100%. All experiments were repeated three times in triplicate.
Statistical analysis
Statistical analyses were performed using SPSS 21.0 software (IBM, USA). Figures were constructed using GraphPad Prism 6.0 software (La Jolla, USA). The results are presented as means ± standard deviations (SD). The statistical significance of differences between groups was analyzed using Student’s t test or the Chi-squared test. The correlations between ADAMTS5 expression and clinicopathological characteristics were evaluated using a Chi-squared test or Fisher’s exact test. OS data were analyzed using the Kaplan–Meier method and compared with a log-rank test. Significant (P < 0.05) factors in the univariate analysis were entered into the multivariate analysis based on the Cox proportional hazards regression model. P < 0.05 was considered statistically significant for all methods.
Results
ADAMTS5 is downregulated in human gastric cancer
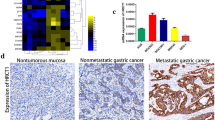
Levels of the ADAMTS5 mRNA in GC samples were measured using the Oncomine microarray database to elucidate the roles of ADAMTS5 in GC initiation and progression. ADAMTS5 was identified as one of the various genes expressed at lower levels in human gastric cancer samples than normal gastric tissue (Fig. 1a). Then, IHC was used to detect the levels of the ADAMTS5 protein in 176 GC samples and 24 adjacent noncancerous tissues. Positive expression of the ADAMTS5 protein was detected in 21 (87.5%) of the 24 noncancerous gastric tissues and 82 (46.6%) of the 176 GC tissues; however, negative expression was observed in only 3 (12.5%) of the normal gastric tissues and 94 (53.4%) of the GC tissues. The ADAMTS5 protein was expressed at significantly lower levels in GC tissues (n = 176; SI = 3.2 ± 3.0) than in noncancerous tissues (n = 24; SI = 7.8 ± 3.2; P < 0.001, Fig. 1b). ADAMTS5 was mainly located in the cytoplasm of normal mucosal epithelial cells and primary cancer cells. Representative images of different ADAMTS5 expression levels in normal gastric tissues and GC tissues are shown in Fig. 1c. Levels of the ADAMTS5 mRNA were also examined in an independent set of 30 pairs of GC tissues matched with adjacent noncancerous tissues by qRT-PCR. Consistent with results from the Oncomine microarray database, the ADAMTS5 mRNA was expressed at lower levels in GC tissues than in noncancerous tissues (Fig. 1d). Meanwhile, the expression of the ADAMTS5 mRNA and protein was also evaluated in 8 gastric cancer cell lines and the normal human gastric epithelial cell line (GES1) using qRT-PCR and western blotting (Fig. 1e). Based on these results, ADAMTS5 is downregulated and involved in gastric cancer.
ADAMTS5 is downregulated in human gastric cancer. a ADAMTS5 mRNA expression in samples from gastric cancer and normal gastric tissue was measured using the Oncomine microarray database. Pooling of 11 analyses from seven different microarray studies shows ADAMTS5 is one of the various genes expressed at lower levels in human gastric cancer samples than normal gastric tissue. P = 0.015, Student’s t test. b ADAMTS5 expression levels in GC tissues by IHC were significantly lower than that of the noncancerous tissues (n = 176, SI = 3.23 ± 2.95 vs. n = 24, SI = 7.79 ± 3.20; P < 0.001). c Representative images of different ADAMTS5 expression levels in normal or GC tissues (magnification ×200 and 400) were shown. d ADAMTS5 mRNA expression was expressed at lower levels in GC tissues than that of corresponding noncancerous tissues (n = 30, P < 0.001) by qRT-PCR assay. e Expression of ADAMTS5 at mRNA and protein levels in 8 gastric cancer cell lines (MGC803, BGC823, AGS, MKN1, SNU1, MKN45, SGC7901, and HGC27) and one immortalized gastric mucosa cell, GES1, were analyzed by qRT-PCR (top panel)and western blotting (bottom panel), respectively. The expression levels of ADAMTS5 mRNA were normalized to that of β-Actin mRNA, which served as a control for the input complementary DNA. The data are expressed as the mean ± standard deviation
Correlations between ADAMTS5 expression and the clinicopathological characteristics and survival of patients with gastric cancer
Subsequently, the correlations between ADAMTS5 expression and the clinicopathological characteristics of patients with GC were explored. ADAMTS5 expression was significantly correlated with gender (P = 0.002), histological type (P = 0.003), degree of differentiation (P < 0.001), M stage (P = 0.001), TNM stage (P = 0.008) and vascular invasion (P = 0.01). In contrast, no statistically significant correlations were observed for age, tumor size, tumor location, T stage or N stage (Table 1). High ADAMTS5 expression was more frequently observed in patients with tubular or papillary adenocarcinoma (77/147, 52.4%) than in patients with signet-ring-cell carcinoma (2/21, 9.5%) or mucinous adenocarcinoma (2/6, 33.3%). Additionally, a significantly greater percentage of patients with well-moderately differentiated GC exhibited high ADAMTS5 expression (34/47, 72.3%) than patients with poorly differentiated GC (48/129, 37.2%). Representative images of IHC staining are shown in Supplementary Figure 1.
The influence of the ADAMTS5 expression status on the OS of patients with GC was also investigated. For all patients in this study, the mean survival time was 78.0 ± 4.2 months, and the 5-year OS rate was 56.6%. The 5-year OS rate for patients with low ADAMTS5 expression was significantly shorter than that for patients with high ADAMTS5 expression (44.7% vs. 69.4%; P = 0.001, Fig. 2a). A further analysis of the 5-year OS rate in patients stratified by TNM stage revealed a significant difference only between the patients with TNM stage III tumors displaying low and high ADAMTS5 expression (40.1% vs. 56.6%; P = 0.01, Fig. 2b) and not patients with TNM stage I (88.9% vs. 100.0%; P = 0.2, Fig. 2b), stage II (69.6% vs. 82.7%; P = 0.2, Fig. 2b) or stage IV tumors (15.5% vs. 50.0%; P = 0.4, Fig. 2b).
Kaplan–Meier survival curves for the GC patients according to ADAMTS5 expression levels. a GC patients with high ADAMTS5 expression had a better overall survival than that of patients with low ADAMTS5 expression (P = 0.007). b For stage by stage analysis, overall survival between patients with high and low ADAMTS5 expression was found to have significant difference at TNM stage III (P = 0.01), not at stage I (P = 0.2), II (P = 0.2) and IV (P = 0.4)
Univariate analyses revealed significant correlations between the tumor invasion depth (P < 0.001), lymph node metastasis (P < 0.001), distant metastasis (P < 0.001) and vascular invasion (P < 0.001) with OS (Table 2). All of these clinicopathological features affecting OS were included in a multivariate Cox proportional hazard regression model to further assess their impacts on the survival of patients with GC. Based on the results obtained using this model, ADAMTS5 (P = 0.04) was confirmed as an independent prognostic factor for patients with GC, as along with invasion depth (P = 0.01) and distant metastasis (P = 0.006) (Table 2).
Transcriptional silencing of ADAMTS5 is associated with promoter methylation in GC cell lines and tissues
As mentioned above, qRT-PCR results revealed the downregulation of ADAMTS5 in GC cell lines and tumor tissues overall, which prompted us to explore the possible mechanisms. We analyzed the role of ADAMTS5 gene promoter methylation in GC cell lines and tumor tissues by methylation-specific PCR. ADAMTS5 gene promoter methylation was not detected in GES1, MGC803, BGC823, MKN1 and SGC7901 cell lines expressing ADAMTS5, but full or partial methylation was detected in AGS, SNU1, MKN45 and HGC27 cell lines (Supplementary Figure 2A). Furthermore, methylation was also detected in gastric cancer tissues. Among five paired tissues, hypermethylation was detected in all five tumor tissues, whereas partial or no methylation was detected in the paired normal tissues (Supplementary Figure 2A). AGS and MKN45 cells that showed silencing of ADAMTS5 were treated with the demethylation agent 5-aza-dC alone or in combination with TSA to determine whether promoter methylation mediated ADAMTS5 silencing. The 5-aza-dC or 5-aza-dC/TSA treatment partially restored the methylated CpG sites to the unmethylated status, and the expression of ADAMTS5 was also restored (Supplementary Figure 2B). Additionally, the level of methylation within the ADAMTS5 promoter region was then characterized and validated by BGS. The BGS results were consistent with methylation-specific PCR (Supplementary Figure 2C and D). Thus, transcriptional silencing of ADAMTS5 in GC samples was associated with promoter methylation.
ADAMTS5 suppresses GC cell migration and invasion
A series of in vivo assays was performed to further elucidate the functions of ADAMTS5 in gastric cancer. Levels of ADAMTS5 in stable overexpressing and knockdown cells were confirmed by qRT-PCR and western blotting (Supplementary Figure 3). ADAMTS5 did not exert significant effects on cell proliferation, the cell cycle and apoptosis (Supplementary Figure 4) in the present study. ADAMTS5 suppressed HUVEC migration by downregulating VEGF expression in a previous study [11], whereas it promoted the migration and invasion of non-small cell lung cancer [15]. Transwell assays and wound healing assays were performed to investigate the roles of ADAMTS5 in GC cell migration and invasion. The transwell migration assays revealed fewer migrated cells in the ADAMTS5 overexpression groups than in the control groups of AGS (241.5 ± 13.8 vs. 371.0 ± 17.5, P < 0.001) and MKN45 cells (204.0 ± 14.0 vs. 415.0 ± 15.4, P < 0.001) (Fig. 3a). Similarly, in the invasion assays, the numbers of cells that crossed the Matrigel-coated membrane in the ADAMTS5 overexpression groups were reduced from 330.8 ± 8.4 to 132.8 ± 11.1 (P < 0.001) for AGS cells and from 305.3 ± 19.0 to 167.0 ± 9.3 (P < 0.001) for MKN45 cells (Fig. 3b) compared with the control groups. Consistent with the results of the transwell assays, ADAMTS5 significantly slowed the wound healing of GC cells compared with the control groups (Fig. 3c). The relative healing of wounded areas in the ADAMTS5 groups at 36 h after scratching was reduced compared with those in the control groups of AGS (50.7 ± 3.1% vs. 100 ± 0.010%, P < 0.001) and MKN45 cells (68.3 ± 3.5% vs. 100 ± 0.0%, P < 0.001). Based on these results, ADAMTS5 upregulation inhibits the migration and invasion of GC cells.
ADAMTS5 suppresses GC cell migration and invasion. a, b Transwell migration and invasion assays showed that ADAMTS5 overexpression suppressed the capabilities of migration and invasion in AGS and MKN45 cells (magnification ×200). c Meanwhile, ADAMTS5 slowed down the wound-healing course of GC cells (magnification ×200). d, e ADAMTS5 knockdown increased significantly the abilities of migration and invasion in SGC7901 and BGC823 cells (magnification ×200), and f accelerated wound-healing course both in SGC7901 and BGC823 cells.*P < 0.001, **P = 0.003
An ADAMTS5-specific shRNA was used to knockdown its expression in SGC7901 and BGC823 cells that expressed endogenous ADAMTS5 protein at high levels to further confirm the inhibitory effect of ADAMTS5 on the malignant phenotype of GC cells (Supplementary Figure 3B). As expected, downregulation of ADAMTS5 significantly promoted the migration and invasion of SGC7901 cells (both P < 0.001, Fig. 3d, e) compared to the control cells. Similar results were also observed in BGC823 cells (both P < 0.001, Fig. 3d, e). Furthermore, ADAMTS5 knockdown also accelerated wound healing in both SGC7901 (P < 0.001, Fig. 3f) and BGC823 (P < 0.05, Fig. 3f) cells when compared with the control groups. Therefore, ADAMTS5 knockdown restores a more aggressive GC cell phenotype.
ADAMTS5 enhances the adhesion of GC cells
Based on the inhibitory effect of ADAMTS5 on migration and invasion, the effect of ADAMTS5 on cell adhesion was also examined. The adhesion assay showed that ADAMTS5 overexpression markedly enhanced the adhesion of AGS (42.8 ± 0.5% vs. 22.7 ± 1.0%, P < 0.001, Fig. 4a) and MKN45 (34.1 ± 2.5% vs. 25.5 ± 1.6%, P < 0.001, Fig. 4a) cells to a fibronectin-coated surface compared to the control groups. In contrast, ADAMTS5 knockdown dramatically inhibited the adhesion of SGC7901 (206.5 ± 7.0 vs. 118.8 ± 4.7, P < 0.001, Fig. 4b) and BGC823 cells (176.8 ± 6.8 vs. 102.8 ± 5.0, P < 0.001, Fig. 4b). Similar results were obtained with the MTS method (both P < 0.001, Fig. 4c). Based on these data, ADAMTS5 inhibits GC cell migration and invasion and simultaneously enhances cell adhesion.
ADAMTS5 enhances the adhesion of GC cells. Cell adhesion assay was performed to examine the abilities of cell adhesion in GC cells with ADAMTS5 overexpression or knockdown. a ADAMTS5 overexpression enhanced the adhesion of AGS and MKN45 cells to ECM, the adherent cells were stained with DAPI and counted under a fluorescence microscopy (magnification ×200). b ADAMTS5 knockdown dramatically decreased the abilities of adhesion in SGC7901 and BGC823 (magnification ×200). c Significant differences were also found in adhesion abilities in cells with ADAMTS5 overexpression or knockdown using the MTS method. *P < 0.001
ADAMTS5 downregulates ETS1 and inhibits angiogenesis in gastric cancer
We analyzed the downstream signaling pathways affected by ADAMTS5 to investigate the underlying mechanisms by which ADAMTS5 regulates GC cell migration and invasion. As ADAMTS5 inhibited the migration and invasion and enhanced the adhesion of GC cells, we examined whether ADAMTS5 regulates the epithelial–mesenchymal transition (EMT), a process that is recognized as a critical event in tumor metastasis [16]. The levels of EMT-related molecules were detected by western blotting. However, ADAMTS5 did not alter the levels of an epithelial marker (E-cadherin), mesenchymal markers (vimentin and N-cadherin), and key EMT-related transcription factors (Snail and Slug) (Supplementary Figure 5), suggesting that ADAMTS5 may not repress the aggressive phenotype of GC cells by regulating EMT.
Based on the anti-angiogenesis properties of ADAMTS5 reported in previous studies [10, 11], the correlation between microvessel density (MVD) and ADAMTS5 expression was determined. MVD was determined by immunohistochemical staining for CD31, a molecular marker of endothelial cells. According to the IHC staining, a negative correlation was observed between MVD and ADAMTS5 expression (r = − 0.3, P < 0.001, Fig. 5a), indicating that ADAMTS5 exerted anti-angiogenesis effects on gastric cancer. Representative images of immunohistochemical staining for ADAMTS5 and CD31 in the same GC tissues are shown in Fig. 5b. Furthermore, the VEGF levels in the supernatant of AGS and MKN45 culture medium from ADAMTS5 overexpression and control groups were detected with an ELISA using methods described in a previous study [10, 11], and no significant difference was observed between the two groups (Supplementary Figure 6). Thus, the anti-angiogenesis effects of ADAMS5 on GC may not be mediated by the VEGF signaling pathway.
ADAMTS5 downregulates ETS1 and inhibits angiogenesis in GC. a The MVD is inversely correlated with ADAMTS5 expression in gastric cancer tissues(r = − 0.3, P < 0.001, n = 176). b Representative immunohistochemical staining images of ADAMTS5, CD31 and ETS1 in the same surgical specimens with high or low expression of ADAMTS5. c ADAMTS5 overexpression significantly downregulated ETS1 expression in both AGS and MKN45 cells, and d ADAMTS5 knockdown upregulated ETS1 expression in both SGC7901 and BGC823 cells determined by qRT-PCR and western blotting, respectively. e Representative examples of negative, weak, moderate, strong staining of ETS1 in GC tissues (magnification ×200 and 400). f Staining index of ETS1 in high ADAMTS5 expression GC tissues was lower than that of low ADAMTS5 expression ones. g MVD in high ETS1 expression GC tissues was greater than that of low ETS1 expression ones. *P < 0.001, **P = 0.048, ***P = 0.04
A MAP Kinase Signal Pathway PCR Array was performed using AGS cells overexpressing ADAMTS5 and the control group. The top 5 upregulated and downregulated genes are listed in Supplementary Table 1. These data indicated a significant downregulation of ETS1 in the ADAMTS5 overexpression group compared with the control group. ETS1 is a transcriptional activator that is involved in various key events involved in cancer progression, EMT and neo-angiogenesis [17]. We next verified the expression of ETS1 in AGS and MKN45 cells. The qRT-PCR and western blot analyses showed that ADAMTS5 overexpression significantly suppressed ETS1 expression at the mRNA and protein levels in both AGS and MKN45 cells (Fig. 5c). In contrast, ADAMTS5 knockdown increased ETS1 expression in both SGC7901 and BGC823 cells (Fig. 5d).
Heterogeneous ETS1 staining was observed in carcinomas and was mainly located in the cytoplasm and nuclei of the tumor cells (Fig. 5e). Among the 176 gastric cancer tissues, 83 showed a high ETS1 expression level of 47.2%. The staining index of ETS1 in the high ADAMTS5 expression group (3.57 ± 3.43) was lower than the low ADAMTS5 expression group (5.23 ± 3.82), and the difference was statistically significant (P = 0.003, Fig. 5f), suggesting a negative correlation between ADAMTS5 and ETS1 expression, consistent with the results obtained from GC cell lines. ETS1 stimulates neo-angiogenesis and thereby promotes tumor growth [17]. We next investigated the correlation between ETS1 expression and MVD in gastric cancer tissues. A greater MVD was observed in tumors expressing high levels of ETS1 than in tumors expressing low levels of ETS1 (73.8 ± 44.7 vs. 61.6 ± 30.8, P = 0.04, Fig. 5g). Thus, the inhibitory effect of ADAMTS5 on angiogenesis might be associated with the downregulation of ETS1.
Discussion
ADAMTS5 belongs to the ADAMTS family, which plays diverse roles in biological and pathological processes [2]. The function of ADAMTS5 varies among different cancer types, as the protein functions as a tumor suppressor in breast cancer [18] and hepatocellular carcinoma [11] and as an oncogene in human glioblastomas and non-small cell lung cancer [13, 15]. These conflicting findings suggest that ADAMTS5 may play a dual role in different pathological types of cancer. However, the functions and mechanisms of ADAMTS5 in GC remain largely unclear. In the present study, the ADAMTS5 mRNA was downregulated in gastric cancer compared to normal tissues in 11 analyses of the Oncomine microarray database. Consistently, we confirmed the downregulation of ADAMTS5 in GC tissues and cell lines. IHC staining revealed correlations between the ADAMTS5 expression status and gender, histological type, degree of differentiation, M stage, TNM stage and vascular invasion. In addition, downregulation of ADAMTS5 was significantly correlated with a poorer OS of patients with GC, particularly in patients with TNM stage III tumors. Because our sample included only a few patients with TNM stage I/II tumors, we observed a trend that high levels of ADAMTS5 in patients with GC correlated with better survival, but the difference was not statistically significant. Therefore, larger numbers of patients with GC are needed to further determine the prognostic value. Nevertheless, ADAMTS5 represents a potential independent prognostic factor for gastric cancer. Furthermore, forced expression of ADAMTS5 in GC cells suppressed migration and invasion in vitro and increased the adhesion of GC cells to ECM. In contrast, ADAMTS5 knockdown partially restored the aggressive phenotype of GC cancer cells. In addition, the negative correlation between ADAMTS5 expression and MVD in gastric cancer tissues suggested that ADAMTS5 exerted anti-angiogenic effects on GC. These findings clearly implicated ADAMTS5 as a tumor suppressor in the initiation and progression of GC, and thus, this protein may be exploited as a prognostic indicator in patients with GC.
ADAMTS5 was transcriptionally silent in GC cells and tissues, as the MSP and BGS results revealed hypermethylation of CpG islands in the ADAMTS5 promoter. Pharmacological demethylation treatments in ADAMTS5-silenced GC cells restored ADAMTS5 expression, indicating that promoter methylation plays a key role in regulating ADAMTS5 expression in gastric cancer. We further analyzed the TCGA database using a Firehose analysis to validate our results. ADAMTS5 was one of the genes displaying a significant negative correlation with gastric cancer, consistent with the results from our present study. Furthermore, a significant negative correlation between the methylation status and expression of the ADAMTS5 mRNA was also identified in 36 of 38 other types of cancer, with the exception of glioblastoma multiforme and ovarian serous cystadenocarcinoma (Supplementary Table 2). We also analyzed mutations (Supplementary Table 3) and single nucleotide polymorphisms (SNPs) (Supplementary Table 4) in the ADAMTS5 gene using the TCGA database but did not identify significant correlations between the expression of the ADAMTS5 mRNA and these two factors. In summary, the epigenetic silencing of ADAMTS5 is involved in some cancer types, including gastric cancer.
Metastasis is a major cause of cancer-related mortality and accounts for approximately 90% of cancer-related deaths [19]. Thus, the mechanism of gastric cancer metastasis must be clarified to develop new therapeutic methods and improve prognoses. HCC cells expressing ADAMTS5 suppress HUVEC migration by downregulating VEGF expression [11]. ADAMTS5 suppressed the migration and invasion and increased the adhesion of GC cells to ECM, suggesting inhibitory effects on the initiation and progression of metastasis. Angiogenesis is an important process associated with both tumor growth and metastasis. This process is regulated by a balance between pro- and anti-angiogenic molecules. Strategies targeting angiogenesis are important methods that are currently used to treat gastric cancer [20, 21]. Several members of the ADAMTS family, including ADAMTS5, have been shown to be involved in regulating angiogenesis [3]. Consistent with the previous reports that ADAMTS5 inhibits angiogenesis in mouse melanoma and human HCC samples [10, 11], ADAMTS5 expression was negatively correlated with MVD in gastric cancer tissues in the present study, which suggested an inhibitory effect of ADAMTS5 on angiogenesis. The suppression of tumor metastasis and angiogenesis by ADAMTS5 suggested a vital role for ADAMTS5 in the development and progression of human gastric cancer. Unlike the reports showing that ADAMTS5 downregulated VEGF [10, 11], VEGF levels in the culture medium of the GC cells were not significantly different between the two groups, suggesting that ADAMTS5 might regulate GC cell migration, invasion and angiogenesis through a VEGF-independent mechanism. A MAP kinase pathway RT-PCR array revealed the downregulation of ETS1 expression by ADAMTS5 in GC cells, which was further confirmed by qRT-PCR and western blot analyses. Consistently, ADAMTS5 expression in GC tissues was negatively correlated with ETS1 expression. Furthermore, MVD was positively correlated with ETS1 expression in GC tissues. Because ETS1 functions as a transcription factor that stimulates cancer-associated neo-angiogenesis, we deduced that ADAMTS5 inhibited angiogenesis, at least in part, by downregulating ETS1. Of course, angiogenesis is a complex process regulated by various factors and is involved in multiple molecular mechanisms. The specific mechanism by which ADAMTS5 regulates tumor angiogenesis requires further study. In addition to inhibiting angiogenesis, ETS1 is also reported to promote invasiveness and induces EMT and drug resistance [17]. However, no evidence supporting the hypothesis that ETS1 enhances the invasion of GC cells by inducing EMT was reported in this study. ADAMTS5 overexpression resulted in the downregulation of ETS1, and its knockdown partially restored ETS1 expression in GC cells. Thus, the effects of ADAMTS5 on migration and invasion of GC cells might be associated with ETS1-related angiogenesis. ETS1 promotes the invasion of cancer cells by upregulating matrix metalloproteinases (MMPs) [22]. Further investigations are needed to determine whether ADAMTS5 suppresses GC cell migration and invasion by downregulating ETS1/MMP(s).
Notably, ADAMTS5 contributed to the migration and invasion of glioblastomas and NSCLC [13, 15], which contradicted our findings in gastric cancer. Dual roles for another member of the ADAMTS family, ADAMTS15, in invasion and metastasis have also been reported. ADAMTS15 promotes breast cancer cell metastasis to the lung but suppresses liver metastasis in a metalloproteinase-dependent and independent manner, respectively [23]. The seemingly opposite effects of ADAMTS15 on metastasis probably depend upon the tissue microenvironment in the metastatic target organ [23]. Researchers have not clearly determined whether the opposite effects of ADAMTS5 on cancer invasion and metastasis in different cancer types are due to different tumor microenvironments or different genotypes. Multiple mechanisms might regulate the effects of ADAMTS5 on invasion and metastasis. Moreover, although ADAMTS5 did not affect GC cell proliferation in vitro, the anti-angiogenesis effect of ADAMTS5 indicated that it inhibits tumor growth, since the nutrients and oxygen supplied by vessels were blocked. The roles of ADAMTS5 in GC cell growth, invasion and angiogenesis in vivo remain unexplored, and thus, an animal model of gastric cancer is needed. The precise mechanism by which ADAMTS5 regulates tumor metastasis and angiogenesis requires further research.
Conclusions
In summary, we demonstrated that ADAMTS5 is downregulated in human gastric cancer, and low expression of ADAMTS5 correlates with a worse overall survival of patients with GC. More importantly, to the best of our knowledge, this study is the first to confirm the biological effects of ADAMTS5 on GC progression, including the suppression of cell migration and invasion, the enhancement of cell adhesion and the inhibition of angiogenesis. A possible mechanism underlying these effects was that ADAMTS5 downregulated ETS1, thus altering its downstream molecules related to tumor metastasis and angiogenesis. These findings provide a basic rationale for the potential use of ADAMTS5 as a prognostic biomarker and therapeutic target for patients with GC. However, the precise mechanism by which ADAMTS5 regulates tumor metastasis and angiogenesis requires further investigation.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 2015;16:113.
Kumar S, Rao N, Ge R. Emerging roles of ADAMTSs in angiogenesis and cancer. Cancers (Basel). 2012;4(4):1252–99.
Cal S, Lopez-Otin C. ADAMTS proteases and cancer. Matrix Biol. 2015;44–46:77–85.
Luque A, Carpizo DR, Iruela-Arispe ML. ADAMTS1/METH1 inhibits endothelial cell proliferation by direct binding and sequestration of VEGF165. J Biol Chem. 2003;278(26):23656–65.
Gustavsson H, Wang W, Jennbacken K, Welen K, Damber JE. ADAMTS1, a putative anti-angiogenic factor, is decreased in human prostate cancer. BJU Int. 2009;104(11):1786–90.
Tan Ide A, Ricciardelli C, Russell DL. The metalloproteinase ADAMTS1: a comprehensive review of its role in tumorigenic and metastatic pathways. Int J Cancer. 2013;133(10):2263–76.
Verma P, Dalal K. ADAMTS-4 and ADAMTS-5: key enzymes in osteoarthritis. J Cell Biochem. 2011;112(12):3507–14.
Kintakas C, McCulloch DR. Emerging roles for ADAMTS5 during development and disease. Matrix Biol. 2011;30(5–6):311–7.
Kumar S, Sharghi-Namini S, Rao N, Ge R. ADAMTS5 functions as an anti-angiogenic and anti-tumorigenic protein independent of its proteoglycanase activity. Am J Pathol. 2012;181(3):1056–68.
Li C, Xiong Y, Yang X, Wang L, Zhang S, Dai N, Li M, Ren T, Yang Y, Zhou SF, et al. Lost expression of ADAMTS5 protein associates with progression and poor prognosis of hepatocellular carcinoma. Drug Des Dev Ther. 2015;9:1773–83.
Kim YH, Lee HC, Kim SY, Yeom YI, Ryu KJ, Min BH, Kim DH, Son HJ, Rhee PL, Kim JJ, et al. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann Surg Oncol. 2011;18(8):2338–47.
Nakada M, Miyamori H, Kita D, Takahashi T, Yamashita J, Sato H, Miura R, Yamaguchi Y, Okada Y. Human glioblastomas overexpress ADAMTS-5 that degrades brevican. Acta Neuropathol. 2005;110(3):239–46.
Huang J, Bai Y, Huo L, Xiao J, Fan X, Yang Z, Chen H, Yang Z. Upregulation of a disintegrin and metalloprotease 8 is associated with progression and prognosis of patients with gastric cancer. Transl Res. 2015;166(6):602–13.
Gu J, Chen J, Feng J, Liu Y, Xue Q, Mao G, Gai L, Lu X, Zhang R, Cheng J, et al. Overexpression of ADAMTS5 can regulate the migration and invasion of non-small cell lung cancer. Tumor Biol. 2016;37(7):8681–9.
Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14(6):818–29.
Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol. 2015;35:20–38.
Porter S, Scott SD, Sassoon EM, Williams MR, Jones JL, Girling AC, Ball RY, Edwards DR. Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin Cancer Res. 2004;10(7):2429–40.
Guan X. Cancer metastases: challenges and opportunities. Acta Pharm Sin B. 2015;5(5):402–18.
Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29(6 Suppl 16):15–8.
Weis SM, Cheresh DA. Tumor angiogenesis: molecular pathways and therapeutic targets. Nat Med. 2017;17(11):1359–70.
Singh S, Barrett J, Sakata K, Tozer RG, Singh G. ETS proteins and MMPs: partners in invasion and metastasis. Curr Drug Targets. 2002;3(5):359–67.
Kelwick R, Wagstaff L, Decock J, Roghi C, Cooley LS, Robinson SD, Arnold H, Gavrilovic J, Jaworski DM, Yamamoto K, et al. Metalloproteinase-dependent and -independent processes contribute to inhibition of breast cancer cell migration, angiogenesis and liver metastasis by a disintegrin and metalloproteinase with thrombospondin motifs-15. Int J Cancer. 2015;136(4):E14–26.
Acknowledgements
The authors sincerely thank Prof. Yan Huang, Xinjuan Fan and Zihuan Yang for providing technical assistance.
Funding
This work was supported by grants from the National Key Clinical Discipline, the National Natural Science Foundation of China (Grant no. 81772594), the Science and Technology Planning Project of Guangdong Province (Grant no. 2014A020212706), the Guangdong Natural Science Foundation (Grant no. 2015A030313008) and the Science and Technology Program of Guangzhou, China (Grant no. 201803010095).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical standards
All procedures were performed in accordance with the ethical standards of the responsible committees on human experimentation (institutional and national) and the 1964 Declaration of Helsinki and its later versions. Informed consent for inclusion in the study, or the equivalent, was obtained from all patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1. Representative images of immunostaining of ADAMTS5 in GC tissues
. (A) High expression of ADAMTS5 was found in tubular adenocarcinoma, and low expression in signet-ring-cell carcinoma and mucinous adenocarcinoma (magnification, ×200 and 400). (B) ADAMTS5 expression status in poorly, moderately, and well differentiated GC tissues; the staining results were negative, moderate and strong, respectively (magnification, ×200 and 400). (TIF 14032 KB)
Supplementary Figure 2. Hypermethylation of ADAMTS5 in gastric cancer. (A)
MSP analysis was carried out in GC cells and tissues using the specific primers for either methylated or unmethylated DNA.T, tumor tissue; N, normal tissue; M, methylated DNA; U, unmethylated DNA, Control 1,2 Genomic DNA from placental tissue treated with SssI methylase or untreated was used as a positive control for methylated DNA or unmethylated DNA, respectively. (B) ADAMTS5 expression was restored in AGS and MKN45 cells after treatment with 5-Aza-dC or 5-Aza-dC/TSA. Methylation status and mRNA expression level of ADAMTS5 were detected by MSP and RT-PCR, respectively. (C) Methylation status of the ADAMTS5 promoter was confirmed by bisulfite genomic sequencing. (D) Chromatogram representative of BGS results in GC cells and tissues. (TIF 9195 KB)
Supplementary Figure 3: ADAMTS5 overexpression or knockdown mediated by lentivirus in GC cells
.(A) ADAMTS5 overexpression in AGS and MKN45 cells and (B) knockdown in SGC7901 and BGC823 cells were confirmed by qRT-PCR and western blotting, respectively. * P < 0.001. (TIF 1094 KB)
Supplementary Figure 4. The effect of ADAMTS5 on cell growth, apoptosis and cell cycle in GC cells
. ADAMTS5 did not affect GC cell proliferation analyzed by RTCA proliferation assay (A) and EdU proliferation assay (B), both P > 0.05. Flow cytometry assay indicated that ADAMTS5 had no effect on GC cells early or late apoptosis(C) and cell cycle arrest(D), both P > 0.05. The experiment was repeated three times in triplicate; the data are expressed as the mean ± standard deviation. (TIF 6623 KB)
Supplementary Figure 5: The effect of ADAMTS5 on the expression of EMT-related proteins in GC cells
. Expression of snail, slug, vimentin, N-cadherin, E-cadherin and β-catenin were detected by western blotting in AGS and MKN45 cells overexpressing ADAMTS5 (A), or SGC7901 and BGC823 cells with ADAMTS5 knockdown (B). There was no markedly difference in expression levels among these EMT-related molecules between ADAMTS5 overexpression/knockdown and control cells. (TIF 2093 KB)
Supplementary Figure 6: The effect of ADAMTS5 on the expression of VEGF in GC cells
. The concentration of VEGF in the supernatant of culture medium was detected by ELISA assay.No significant difference was found in AGS and MKN45 cells between ADAMTS5 overexpression and control groups.*P > 0.05. (TIF 22 KB)
Rights and permissions
About this article
Cite this article
Huang, J., Sun, Y., Chen, H. et al. ADAMTS5 acts as a tumor suppressor by inhibiting migration, invasion and angiogenesis in human gastric cancer. Gastric Cancer 22, 287–301 (2019). https://doi.org/10.1007/s10120-018-0866-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-018-0866-2