Abstract
Background
Transcribed ultraconserved regions (T-UCRs) are a novel class of noncoding RNAs that are highly conserved among the orthologous regions in most vertebrates. It has been reported that T-UCRs have distinct signatures in human cancers. We previously discovered the downregulation of T-UCR expression in gastric cancer (GC), indicating that T-UCRs could play an important role in GC biology. Uc.160+, a T-UCR reported to be downregulated in human cancer, has not been examined in GC.
Methods
We analyzed the expression pattern of Uc.160+ in nonneoplastic and tumor tissues of the stomach by using uantitative reverse transcription polymerase chain reaction (qRT-PCR) and in situ hybridization (ISH), specifically focusing on the mechanism of transcriptional regulation and target genes that are regulated by T-UCRs. We also attempted to determine the effect of Uc.160+ expression on biological features of GC cell lines by Western blotting.
Results
On the basis of the qRT-PCR and ISH results, Uc.160+ expression in adenoma and GC tissues was clearly downregulated compared with that in nonneoplastic mucosa tissues of the stomach. Cancer-specific DNA methylation in the promoter region of Uc.160 was observed by bisulfite genomic DNA sequencing analysis. The effect of DNA methylation on Uc.160+ expression was further confirmed by reporter gene assay. We also revealed that Uc.160+ inhibited the phosphorylation of Akt by regulating phosphatase and tensin homolog (PTEN) expression.
Conclusions
These results indicate that Uc.160+ could possibly have a tumor suppressive role in GC.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is the fifth leading cause of global cancer-related death and the most frequent cancer in East Asian countries. Despite improvements in diagnostic and therapeutic methods, GC still causes more than 840,000 deaths a year [1]. GC is thought to originate from a sequential accumulation of genetic and epigenetic alterations in stomach epithelial cells. An in-depth investigation of the molecular biology of GC is necessary to further elucidate its molecular mechanism.
Transcribed ultraconserved regions (T-UCRs) are a novel class of long noncoding RNAs transcribed from ultraconserved regions (UCRs). They are located in both intragenic and intergenic regions and are highly conserved among orthologous regions of vertebrate genomes. There are at least 481 UCRs [2], and these regions can produce 962 T-UCRs: one of the T-UCRs produced by each UCR corresponds to the sense genomic sequence (named “+”) and the other corresponds to the complementary sequence (named “+A”). Recent studies revealed two major molecular mechanisms in the regulation of T-UCR expression: interaction with microRNAs (miRNAs) [3] and CpG island hypermethylation [4]. Most of the T-UCRs found in normal human tissues are ubiquitous, tissue specific, or both [4].
Uc.160 is a UCR on chromosomes band 5q14.1 that is classified as nonexonic (without evidence of an encoding protein). Uc.160 has a CpG island upstream of its coding region, and cancer-specific hypermethylation of this region is observed in various types of cancer cell lines. Furthermore, the methylation frequency of its CpG island was shown to be significantly correlated with tumor metastasis [4]. Among all human cancers, GC is one of the well-established cancers that are closely related to epigenetic alterations rather than genetic changes [5]. It is also well known that the aberrant DNA methylation of various specific genes is induced by exposure to chronic inflammation triggered by Helicobacter pylori [6, 7], which almost exclusively causes GC. Recently, several studies have further confirmed that certain GCs can exhibit a CpG island methylator phenotype, characterized by genome-wide methylation of CpG islands as revealed by next-generation sequencing [8,9,10]. Although CpG island hypermethylation is not limited to Uc.160+, one study demonstrated the correlation between T-UCRs and GC [11]; thus, the examination of Uc.160 in GC could potentially provide novel insights into its molecular machinery.
In this study, to confirm the expression pattern of Uc.160+ in gastric tumor tissues, we evaluated Uc.160+ expression in gastric tumor samples by in situ hybridization (ISH), quantitative reverse transcription polymerase chain reaction (qRT-PCR), and DNA methylation analysis. We also attempted to investigate the methylation status of the CpG island upstream (right) of the Uc.160 coding region and the promoter activity of the CpG island during Uc.160 translation in GC. Furthermore, we revealed the interaction between Uc.160+ and representative molecular pathways by Western blot analysis.
Materials and methods
Cell lines and drug treatment
Four GC cell lines of the MKN series and CRC cell line (DLD-1) were purchased from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). All cell lines were maintained in RPMI-1640 (Nissui Pharmaceutical, Tokyo, Japan) containing 10% fetal bovine serum (BioWhittaker, Walkersville, MD, USA), 2 mM l-glutamine, 50 U/mL penicillin, and 50 μg/mL streptomycin in a humidified atmosphere of 5% CO2 and 95% air at 37 °C. Cells were treated with 1 μM 5-aza-2′-deoxycytidine (Sigma Chemical, St Louis, MO, USA) for 48 h to achieve demethylation.
Tissue samples
For qRT-PCR and bisulfite genomic DNA sequencing, 51 GC samples and corresponding nonneoplastic gastric mucosa samples were used. The samples were frozen immediately in liquid nitrogen and stored at −80 °C until use. The A 260/A 280 ratios of each sample are given in Table S1. The samples were randomly selected from patients who underwent surgery at Hiroshima University Hospital or an affiliated hospital in the years 2005–2008. Comprehensive approvals for basic or clinical research were obtained from all of the patients. This study was conducted in accordance with the Ethical Guidance for Human Genome/Gene Research of the Japanese Government and Human Genome Research of Hiroshima University.
Quantitative reverse transcription polymerase chain reaction analysis
Total RNA was isolated from frozen samples or cancer cell lines with use of Isogen (Nippon Gene, Tokyo, Japan), and 1 µg of total RNA was converted to complementary DNA with a first-strand complementary DNA synthesis kit (Amersham Biosciences, Piscataway, NJ, USA). The primers used are listed in Table 1. Real-time detection of the emission intensity of SYBR Green bound to double-stranded DNA was performed with a CFX Connect real-time system (Bio-Rad Laboratories, Hercules, CA, USA). ACTB-specific polymerase chain reaction products, which were amplified from the same RNA samples, served as internal controls. A single peak on the melting curve analysis was confirmed for each reaction. Relative quantification was determined by the ∆∆Ct method.
Genomic DNA extraction and bisulfite genomic DNA sequencing
Genomic DNAs were extracted with a genomic DNA purification kit (Promega, Madison, WI, USA). To examine DNA methylation patterns, genomic DNAs were treated with 3 M sodium bisulfite as described previously [12, 13]. For analysis of the DNA methylation of Uc.160+, we performed bisulfite genomic sequencing analysis. To obtain products for sequencing, polymerase chain reaction was performed with each 50-μL reaction mixture containing 0.2 μm deoxynucleotide triphosphates, 10 mM tris(hydroxymethyl)aminomethane–HCl (pH 8.3), 50 mm KCl, 2 mm MgCl2, 0.3 μm primers, and 0.75 U AmpliTaq Gold (Applied Biosystems, Foster City, CA, USA). The primers used are listed in Table 1. Individual cloned fragments were sequenced with an ABI PRISM 310 genetic analyzer (Applied Biosystems) as described previously [14].
Transient transfection and luciferase assay
The genomic sequence of Uc.160+ upstream was cloned into the pGL3 basic vector (Promega) with use of its multiple cloning site (160+Luc). MKN-1 or MKN-45 cells (1.0 × 104) were transfected with the construct in a six-well culture dish with FuGENE6 (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer’s instructions. After 48 h, cells were harvested with a dual luciferase reporter assay kit (Promega), and luciferase activity was measured with a GloMax luminometer (Promega) as described previously [15, 16].
In vitro methylation assay
In vitro methylation assays were performed according to previously described methods. Briefly, 1 µg plasmid DNA containing the Uc.160+ promoter construct was incubated for 2 h with 2 U SssI methylase (New England BioLabs, Ipswich, UK). After phenol extraction and ethanol precipitation, MKN-1 or MKN-45 cells were transiently transfected with equal amounts (400 ng) of methylated or unmethylated reporter constructs. Luciferase activity was examined as described earlier.
In situ hybridization
For visualization of Uc.160+, ISH of Uc.160+ was performed as described previously [17]. Briefly, a biotin 3′-labeled locked nucleic acid incorporated probe (custom locked nucleic acid messenger RNA detection probe, Exiqon, Woburn, MA, USA) was used (Table 1). The formalin-fixed, paraffin-embedded tissue samples were sectioned, deparaffinized, and rehydrated. Sample slides were treated with proteinase K (Dako Cytomation, Carpinteria, CA, USA) for 20 min. A 20 nM probe was hybridized with 1× Enzo ISH buffer (Exiqon). The sample slides were heated to 60 °C for 5 min and incubated at 37 °C for 14 h. Immunological detection was performed with a Dako GenPoint tyramide signal amplification system for biotin-labeled probes (Dako Cytomation). The sections were then exposed to a streptavidin–peroxidase reaction and developed with 3,3′-diaminobenzidine.
Uc.160+ expression vector and Western blot analysis
The genomic sequence of Uc.160+ was cloned into the pcDNA3.1 vector (Promega) at the KpnI and XhoI sites as described previously [18]. MKN-1 cells (8.0 × 105) or MKN-45 cells (2.0 × 106) were transfected with the construct in a culture dish with FuGENE6 (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer’s instructions.
For Western blot analysis, cells were lysed as previously described [19, 20]. The lysates were solubilized in Laemmli sample buffer by boiling and subjected to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, followed by electrotransfer onto a nitrocellulose membrane. The membrane was incubated with the primary antibodies listed in Table 2. Immunocomplexes were visualized with an ECL Prime Western blot detection system (GE Healthcare, Little Chalfont, UK). Protein levels were quantified with use of ImageJ version 1.51d (National Institutes of Health, Bethesda, MD, USA) after densitometric scanning of the films.
RNA interference and cell growth assay
Silencer Select small interfering RNA (siRNA; Ambion, Austin, TX, USA) against Uc.160+ was used for RNA interference. Three independent oligonucleotides and negative control siRNA (Invitrogen) were used. The sequence of siRNA no. 1 was 5′-GCACUAUUGCAAGAGCAUUTT-3′, and that of siRNA no. 2 was 5′-GAAAUGAGAUGCACAUUUATT-5′. A total of 1.0 × 106 DLD-1 cells were plated on a 10-cm culture dish 24 h before transfection. Transfection was performed with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Cells were used 48 h after transfection in each of the experiments and assays. To examine cell growth, a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay was performed as described previously [21].
Statistical analysis
Statistical differences were evaluated by a two-tailed Student’s t test or the Mann–Whitney U test. The correlation between expression levels of Uc.160+ and clinicopathological characteristics was analyzed with Fisher’s exact test. The results are expressed as the mean ± standard deviation of triplicate measurements. We considered p < 0.05 to be statistically significant.
Results
Suppression of Uc.160+ expression in gastric adenoma and adenocarcinoma
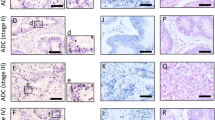
We first attempted to determine the expression of Uc.160+ in nonneoplastic and tumorous tissues of the stomach by ISH. Robust Uc.160+ expression was detected in nonneoplastic tissues of the stomach, including the fundic glands (Fig. 1a) and the glands displaying intestinal metaplasia (Fig. 1b). In contrast, Uc.160+ expression was rather weak in the tubular adenoma (Fig. 1c), and there was little to no expression in certain areas of GC tissues, such as the apical part of well-differentiated tubular adenocarcinoma (Fig. 1d), diffuse-type poorly differentiated adenocarcinoma (Fig. 1e), and the invasive front of tubular adenocarcinoma (Fig. 1f). We then performed qRT-PCR analysis on both GC and nonneoplastic mucosa tissues of all 51 samples, and found that Uc.160+ expression in GC tissues was significantly downregulated (p = 0.0034) compared with that in nonneoplastic mucosa tissues (Fig. 1g). Additional studies focusing on the correlation between expression levels of Uc.160 and clinicopathological characteristics did not show any significance (Table 3). These data suggest that the loss of Uc.160+ could play a key role in tumorigenesis of the stomach.
Uc.160+ expression in nonneoplastic stomach mucosa, tubular adenoma, and gastric cancer (GC) tissues. In situ hybridization analysis of Uc.160+ in the a fundic gland, b intestinal metaplasia of the stomach, c tubular adenoma, d intestinal-type GC, e diffuse-type GC, and f the invasive region of GC. Insets show higher-powered views of each tissue. Magnification: a, b ×100; a, b inserts and c–f ×400. g Quantitative reverse transcription polymerase chain reaction analysis of Uc.160+ in GC and nonneoplastic mucosa tissues
Suppression of Uc.160+ expression by DNA hypermethylation
Although our data indicate that loss of Uc.160+ expression is generally observed in GC tissue, the precise mechanism that underlies Uc.160+ repression remains to be determined. We performed qRT-PCR analysis in four GC cell lines and two normal stomach tissue samples. Uc.160+ was downregulated in all of the four GC cell lines compared with the normal stomach samples (Fig. 2a), which was consistent with the ISH results. Next, we evaluated the expression levels of Uc.160+ in GC cells treated with 5-aza-2′-deoxycytidine to investigate whether these regions were suppressed by DNA-methylation-associated inactivation. In both MKN-1 and MKN-45 cells, Uc.160+ levels were restored by demethylation (Fig. 2b). To assess the extent of Uc.160+ methylation in GC, we searched for and identified CpG islands upstream of Uc.160+ (Fig. 2c).
CpG island methylation of Uc.160+ in gastric cancer (GC) cells. a Uc.160+ expression level in normal stomach and GC cells. b Uc.160+ expression in MKN-1 and MKN-45 cells treated with 5-aza-2′-deoxycytidine (aza-dC). c The sequence upstream of Uc.160+. The vertical lines indicate the positions of CpG dinucleotides. d Bisulfite genomic DNA sequencing analysis of Uc.160+ in GC cells, normal stomach cells, GC cells treated with aza-dC, and GC tissues. Eight single clones represent each sample. A methylated or unmethylated cytosine is indicated by a black square or a white square respectively. The requencing range is indicated by the black arrow in c. e Design of the luciferase vector containing the CpG island upstream of Uc.160+ (160+Luc) and 160+Luc treated with Sssl methylase (160+Met). f Luciferase activity of MKN-1 and MKN-45 cells transfected with a control reporter vector, a reporter vector containing 160+Luc or 160+Met
We next performed bisulfite genomic sequencing and found dense DNA methylation in the upstream region of Uc.160+ in GC cell lines and two GC tissue samples; in contrast, this region was far less methylated in normal stomach tissue samples. We also confirmed that some of the CpG islands in MKN-45 cells treated with 5-aza-2′-deoxycytidine were demethylated (Fig. 2d).
For further investigation of the promoter activity of this region during Uc.160 translation, we constructed a luciferase vector containing the CpG island upstream of Uc.160+ (160+Luc) and an in vitro methylated vector of 160+Luc treated with Sssl methylase (160+Met) (Fig. 2e). When the Uc.160+ upstream sequence was methylated, reporter activity was significantly suppressed (Fig. 2f). These results imply that Uc.160+ expression was tightly regulated by DNA methylation of its promoter region upstream (right) of the coding region.
Inhibition of Akt phosphorylation by Uc.160+
Previous studies have demonstrated that Uc.160+ is methylated in several cancers; however, there are no reports on the biological function of Uc.160+. To elucidate the actual function of Uc.160+ in cancer biology, we first constructed a vector containing the whole sequence of the Uc.160+ coding region and transfected MKN-1 and MKN-45 cells with it. As shown in Fig. 3a, transfection of the vector significantly induced Uc.160+ overexpression. Next, we performed Western blot analysis for the molecules that are involved in the phosphatidylinositol 3-kinase–mitogen-activated protein kinase (MAPK) signaling pathway, pAkt and phosphatase and tensin homolog (PTEN) (Fig. 3b). Although extracellular-signal-regulated kinase, phosphorylated extracellular-signal-regulated kinase, and Akt expression did not change even after Uc.160+ overexpression, the expression of phosphorylated Akt was dramatically decreased, and PTEN expression was inversely increased. These results suggest that Uc.160+ exclusively repressed the phosphorylation of Akt by regulating PTEN expression.
Inhibition of Akt phosphorylation by Uc.160+ through phosphatase and tensin homolog (PTEN) expression regulation. a Quantitative reverse transcription polymerase chain reaction analysis of Uc.160+ expression in MKN-1 and MKN-45 cells transfected with an empty (control) vector or Uc.160+ vector. b Western blot analysis for extracellular-signal-regulated kinase (ERK), phosphorylated ERK (pERK), Akt, phosphorylated Akt (pAkt), and PTEN using MKN-1 and MKN-45 cells transfected with an empty (control) vector or Uc.160+ vector. β-Actin served as a loading control
Effect of deregulation of Uc.160+ expression on cancer cell growth
To investigate the biological significance of Uc.160+ in cancer cells, we performed a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay 4 days after altering Uc.160+ expression. We initially intended to use GC cell lines with transient Uc.160+ overexpression for this study; however, we could not obtain GC cells with sufficient cell growth after Uc.160+ restoration (data not shown). In this experiment, we used DLD-1 cells transfected with Uc.160+-specific siRNAs because DLD-1 possessed the highest endogenous Uc.160+ expression among colorectal cancer cell lines (Fig. S1a). Successful suppression of Uc.160+ in siRNA-treated cells was confirmed by qRT-PCR (Fig. S1b), and growth of DLD-1 cells with downregulated Uc.160+ was significantly reduced compared with that of the negative control siRNA-transfected DLD-1 cells (Fig. S1c). These results indicated that Uc.160+ could be involved in cancer cell growth through regulation of PTEN and potentially other cancer-related genes.
Discussion
Recent studies strongly support the involvement of noncoding RNAs, such as miRNAs and T-UCRs, in the pathogenesis of most cancers investigated and add a new layer of complexity to the molecular architecture of human cancers [22, 23]. Although more detailed studies on miRNAs have been conducted, characterizing the functional significance of T-UCRs in human cancers is also important as a myriad of putative functions of T-UCRs can be hypothesized, including an antisense inhibitory role for protein-coding genes or other noncoding RNAs [24, 25]. We have described here the involvement of Uc.160+ in the cancer biology of GC and revealed the marked silencing of Uc.160+ expression in GC by methylation of the promoter region, which led to the activation of MAPK signaling via repression of PTEN expression. To our knowledge, our study is the first to demonstrate the direct interaction between T-UCR and an essential component of the representative signaling pathway in cancer biology.
Although the precise mechanism of stepwise carcinogenesis in GC, such as whether intestinal metaplasia and gastric adenoma are truly premalignant regions of GC, is still controversial, accumulating evidence indicates that aberrant CpG island hypermethylation of tumor suppressor genes tends to accumulate with the multistep carcinogenesis in GC [26, 27]. Consistent with these results, Uc.160+ expression gradually decreased in accordance with the histological shift from intestinal metaplasia to GC, and the Uc.160 promoter region in GC tissues and cell lines was considerably methylated compared with that in normal stomach tissues. H. pylori infection has been demonstrated as a major risk factor for GC carcinogenesis that contributes to the inactivation of a series of tumor suppressor genes, including RUNX3 [27], CDKN2A [28], TFF2 [29], CDH1 [30], by inducing the methylation of the promoter region of these genes. Taken together, downregulation of Uc.160+ by the methylation of its promoter region could also contribute to GC carcinogenesis. One of the plausible mechanisms that Uc.160+ could be involved in is activation of the MAPK signaling pathway, which leads to runt-related transcription factor 3 (RUNX3) inactivation [31]. These data strongly suggest that Uc.160 contributes to the carcinogenesis in GC by repressing PTEN expression.
We suggest that Uc.160+ is mainly regulated by epigenetic machinery on its promoter region; however, we suspect that Uc.160 may interact with certain miRNAs, and the interaction could play a role in the regulation of PTEN expression. By using miRBase (http://www.mirbase.org/) to identify miRNAs that have consensus sequences with Uc.160, we found candidate miRNAs, such as miR-186-3p, miR-126-5p, and miR-4299. We then validated the target genes of these miRNAs through TargetScanHuman (http://www.targetscan.org/) and revealed that miR-4299 could potentially regulate PTEN expression. This finding demonstrates the complicated network between T-UCRs and miRNAs. Additional studies that use state-of-the-art techniques, such as next-generation sequencing and in silico analyses, will likely be needed to advance our understanding of the involvement of T-UCRs and miRNAs in GC progression.
In conclusion, we have demonstrated the important roles of Uc.160+ in GC carcinogenesis, specifically the regulatory machinery of Uc.160+ expression and its direct interaction with one of the vital signaling pathways in cancer biology. We believe that this study is an important step toward effectively identifying T-UCRs that have an essential function in cancer biology. Our findings call for more studies on the interactions between T-UCRs and miRNAs in cancer biology.
References
Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, et al. The global burden of cancer 2013. JAMA Oncol. 2015;1(4):505–27.
Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–5.
Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–29.
Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–401.
Ushijima T, Sasako M. Focus on gastric cancer. Cancer Cell. 2004;5:121–5.
Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124:2367–74.
Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430–40.
Matsusaka K, Kaneda A, Nagae G, Ushiku T, Kikuchi Y, Hino R, et al. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71:7187–97.
Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K, et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438–42.
Zouridis H, Deng N, Ivanova T, Zhu Y, Wong B, Huang D, et al. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci Transl Med. 2012;4:156ra40.
Goto K, Ishikawa S, Honma R, Tanimoto K, Sakamoto N, Sentani K, et al. The transcribed-ultraconserved regions in prostate and gastric cancer: DNA hypermethylation and microRNA-associated regulation. Oncogene. 2016;27:3598–606.
Hamai Y, Oue N, Mitani Y, Nakayama H, Ito R, Matsusaki K, et al. DNA hypermethylation and histone hypoacetylation of the HLTF gene are associated with reduced expression in gastric carcinoma. Cancer Sci. 2003;94:692–8.
Oue N, Oshimo Y, Nakayama H, Ito R, Yoshida K, Matsusaki K, et al. DNA methylation of multiple genes in gastric carcinoma: association with histological type and CpG island methylator phenotype. Cancer Sci. 2003;94:901–5.
Matsumura S, Oue N, Mitani Y, Kitadai Y, Yasui W. DNA demethylation of vascular endothelial growth factor-C is associated with gene expression and its possible involvement of lymphangiogenesis in gastric cancer. Int J Cancer. 2007;120:1689–95.
Naito Y, Oue N, Hinoi T, Sakamoto N, Sentani K, Ohdan H, et al. Reg IV is a direct target of intestinal transcriptional factor CDX2 in gastric cancer. PLoS ONE. 2012;7:e47545.
Sakamoto N, Oue N, Sentani K, Anami K, Uraoka N, Naito Y, et al. Liver-intestine cadherin induction by epidermal growth factor receptor is associated with intestinal differentiation of gastric cancer. Cancer Sci. 2012;103:1744–50.
Sakamoto N, Naito Y, Oue N, Sentani K, Uraoka N, Zarni Oo H, et al. MicroRNA-148a is downregulated in gastric cancer, targets MMP7, and indicates tumor invasiveness and poor prognosis. Cancer Sci. 2014;105:236–43.
Oo HZ, Sentani K, Sakamoto N, Anami K, Naito Y, Uraoka N, et al. Overexpression of ZDHHC14 promotes migration and invasion of scirrhous type gastric cancer. Oncol Rep. 2014;32:403–10.
Hayashi K, Yokozaki H, Goodison S, Oue N, Suzuki T, Lotan R, et al. Inactivation of retinoic acid receptor beta by promoter CpG hypermethylation in gastric cancer. Differentiation. 2001;68:13–21.
Yasui W, Ayhan A, Kitadai Y, Nishimura K, Yokozaki H, Ito H, et al. Increased expression of p34cdc2 and its kinase activity in human gastric and colonic carcinomas. Int J Cancer. 1993;53:36–41.
Oue N, Naito Y, Hayashi T, Takigahira M, Kawano-Nagatsuma A, Sentani K, et al. Signal peptidase complex 18, encoded by SEC11A, contributes to progression via TGF-alpha secretion in gastric cancer. Oncogene. 2014;33:3918–26.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66.
Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69.
Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413.
Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502.
Kang GH, Lee S, Kim JS, Jung HY. Profile of aberrant CpG island methylation along multistep gastric carcinogenesis. Lab Invest. 2003;83:519–26.
Lu XX, Yu JL, Ying LS, Han J, Wang S, Yu QM, et al. Stepwise cumulation of RUNX3 methylation mediated by Helicobacter pylori infection contributes to gastric carcinoma progression. Cancer. 2012;118:5507–17.
Neves Filho EH, Alves MK, Lima VP, Rabenhorst SH. MTHFR C677T polymorphism and differential methylation status in gastric cancer: an association with Helicobacter pylori infection. Virchows Arch. 2010;457:627–33.
Peterson AJ, Menheniott TR, O’Connor L, Walduck AK, Fox JG, Kawakami K, et al. Helicobacter pylori infection promotes methylation and silencing of trefoil factor 2, leading to gastric tumor development in mice and humans. Gastroenterology. 2010;139:2005–17.
Ferrasi AC. Helicobacter pyloriand EBV in gastric carcinomas: methylation status and microsatellite instability. World J Gastroenterol. 2010;16:312.
Liu Z, Xu X, Chen L, Li W, Sun Y, Zeng J, et al. Helicobacter pylori CagA inhibits the expression of Runx3 via Src/MEK/ERK and p38 MAPK pathways in gastric epithelial cell. J Cell Biochem. 2012;113:1080–6.
Acknowledgements
We thank Shinichi Norimura for excellent technical assistance. We thank the Analysis Center of Life Science, Hiroshima University, for the use of its facilities. This work was supported by Grants-in-Aid for Scientific Research (JP15H04713, JP16K08691) and Grants-in-Aid for Challenging Exploratory Research (26670175, JP16K15247) from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human rights statement and informed consent
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions. Informed consent or substitute for it was obtained from all patients for their being included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10120_2017_714_MOESM1_ESM.tif
Fig. S1 Functional analysis using Uc.160+-silenced cancer cells. a Uc.160+ expression in normal colon and colon cancer cells. b Uc.160+ expression in DLD-1 cells transfected with negative control or two different small interfering RNAs. c The growth-promotive effect of Uc.160+ knockout in DLD-1 cells. The results are expressed as the mean ± standard deviation of triplicate measurements (TIFF 562 kb).
Rights and permissions
About this article
Cite this article
Honma, R., Goto, K., Sakamoto, N. et al. Expression and function of Uc.160+, a transcribed ultraconserved region, in gastric cancer. Gastric Cancer 20, 960–969 (2017). https://doi.org/10.1007/s10120-017-0714-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-017-0714-9