Abstract
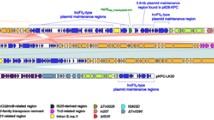
The aim of this review is to provide an update on the plasmids mediating DHA-1 cephalosporinase in Klebsiella pneumoniae. These plasmids have been mainly found in this bacterium but not only. The first was isolated from Salmonella sp. in France in the early 1990s. They are currently reported worldwide. BlaDHA-1 beta-lactamase gene is usually co-expressed with many other antibiotic resistance genes such as extended-spectrum β-lactamases (blaCTX-M-, bla SHV -types), oxacillinases (blaOXA-1, blaOXA-30), penicillinases (bla TEM -type), carbapenemases (bla OXA48 , blaKPC-2), aminoglycosides (aacA, aadA, armA), fluoroquinolones (qnrB4, aac6′-1b-cr), and sulfonamide (sul1) resistance genes. Plasmids carrying DHA-1 cephalosporinase have different sizes (22 to 313 kb), belong to diverse groups of incompatibility (R, L/M, FII(k), FIB, A/C2, HI2, HIB), and are self-transferable or not. The multidrug resistance region consists of a mosaic structure composed of resistance genes, insertion sequences, composite transposon, and integrons.



Similar content being viewed by others
References
Jacoby GA (2009) AmpC beta-lactamases. Clin Microbiol Rev 22:161–182, Table of Contents. https://doi.org/10.1128/CMR.00036-08
Papanicolaou GA, Medeiros AA, Jacoby GA (1990) Novel plasmid-mediated beta-lactamase (MIR-1) conferring resistance to oxyimino- and alpha-methoxy beta-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother 34:2200–2209
Philippon A, Arlet G, Jacoby GA (2002) Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother 46:1–11
Gaillot O, Clément C, Simonet M, Philippon A (1997) Novel transferable beta-lactam resistance with cephalosporinase characteristics in Salmonella enteritidis. J Antimicrob Chemother 39:85–87
Ambler RP (1980) The structure of beta-lactamases. Philos Trans R Soc Lond Ser B Biol Sci 289:321–331
Bush K, Jacoby GA, Medeiros AA (1995) A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother 39:1211–1233
Nourrisson C, Tan RN, Hennequin C, Gibold L, Bonnet R, Robin F (2015) The MAST® D68C test: an interesting tool for detecting extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol 34:975–983. https://doi.org/10.1007/s10096-014-2305-6
Maraskolhe DL, Deotale VS, Mendiratta DK, Narang P (2014) Comparision of three laboratory tests for detection of AmpC β lactamases in Klebsiella species and E. coli. J Clin Diagn Res JCDR 8:DC05–DC08. https://doi.org/10.7860/JCDR/2014/8256.4432
Reuland EA, Hays JP, de Jongh DMC, Abdelrehim E, Willemsen I, Kluytmans JAJW et al (2014) Detection and occurrence of plasmid-mediated AmpC in highly resistant gram-negative rods. PLoS One 9:e91396. https://doi.org/10.1371/journal.pone.0091396
Edquist P, Ringman M, Liljequist BO, Wisell KT, Giske CG (2013) Phenotypic detection of plasmid-acquired AmpC in Escherichia coli—evaluation of screening criteria and performance of two commercial methods for the phenotypic confirmation of AmpC production. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol 32:1205–1210. https://doi.org/10.1007/s10096-013-1869-x
Pérez-Pérez FJ, Hanson ND (2002) Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40:2153–2162
Alvarez M, Tran JH, Chow N, Jacoby GA (2004) Epidemiology of conjugative plasmid-mediated AmpC beta-lactamases in the United States. Antimicrob Agents Chemother 48:533–537
Moland ES, Black JA, Ourada J, Reisbig MD, Hanson ND, Thomson KS (2002) Occurrence of newer beta-lactamases in Klebsiella pneumoniae isolates from 24 U.S. hospitals. Antimicrob Agents Chemother 46:3837–3842
Liebana E, Batchelor M, Clifton-Hadley FA, Davies RH, Hopkins KL, Threlfall EJ (2004) First report of Salmonella isolates with the DHA-1 Amp. beta-lactamase in the United Kingdom. Antimicrob Agents Chemother 48:4492. https://doi.org/10.1128/AAC.48.11.4492.2004
Verdet C, Benzerara Y, Gautier V, Adam O, Ould-Hocine Z, Arlet G (2006) Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob Agents Chemother 50:607–617. https://doi.org/10.1128/AAC.50.2.607-617.2006
Pai H, Kang C-I, Byeon J-H, Lee K-D, Park WB, Kim H-B et al (2004) Epidemiology and clinical features of bloodstream infections caused by AmpC-type-beta-lactamase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 48:3720–3728. https://doi.org/10.1128/AAC.48.10.3720-3728.2004
Pai H, Seo M-R, Choi TY (2007) Association of QnrB determinants and production of extended-spectrum beta-lactamases or plasmid-mediated AmpC beta-lactamases in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother 51:366–368. https://doi.org/10.1128/AAC.00841-06
Yan J-J, Ko W-C, Jung Y-C, Chuang C-L, Wu J-J (2002) Emergence of Klebsiella pneumoniae isolates producing inducible DHA-1 beta-lactamase in a university hospital in Taiwan. J Clin Microbiol 40:3121–3126
Kim J-Y, Park Y-J, Lee S-O, Song W, Jeong SH, Yoo YA et al (2004) Case report: bacteremia due to Salmonella enterica serotype Montevideo producing plasmid-mediated AmpC beta-lactamase (DHA-1). Ann Clin Lab Sci 34:214–217
Yan J-J, Ko W-C, Wu H-M, Tsai S-H, Chuang C-L, Wu J-J (2004) Complexity of Klebsiella pneumoniae isolates resistant to both cephamycins and extended-spectrum cephalosporins at a teaching hospital in Taiwan. J Clin Microbiol 42:5337–5340. https://doi.org/10.1128/JCM.42.11.5337-5340.2004
Roh KH, Uh Y, Kim J-S, Kim H-S, Shin DH, Song W (2008) First outbreak of multidrug-resistant Klebsiella pneumoniae producing both SHV-12-type extended-spectrum beta-lactamase and DHA-1-type AmpC beta-lactamase at a Korean hospital. Yonsei Med J 49:53–57. https://doi.org/10.3349/ymj.2008.49.1.53
Tamang MD, Seol SY, Oh J-Y, Kang HY, Lee JC, Lee YC et al (2008) Plasmid-mediated quinolone resistance determinants qnrA, qnrB, and qnrS among clinical isolates of Enterobacteriaceae in a Korean hospital. Antimicrob Agents Chemother 52:4159–4162. https://doi.org/10.1128/AAC.01633-07
Hu F, Xu X, Zhu D, Wang M (2008) Coexistence of qnrB4 and qnrS1 in a clinical strain of Klebsiella pneumoniae. Acta Pharmacol Sin 29:320–324. https://doi.org/10.1111/j.1745-7254.2008.00757.x
Lee C-H, Liu J-W, Li C-C, Chien C-C, Tang Y-F, Su L-H (2011) Spread of ISCR1 elements containing bla DHA- 1 and multiple antimicrobial resistance genes leading to increase of flomoxef resistance in extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 55:4058–4063. https://doi.org/10.1128/AAC.00259-11
Jiang Y, Yu D, Wei Z, Shen P, Zhou Z, Yu Y (2010) Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying bla KPC-2 , bla DHA-1 , qnrB4, and armA. Antimicrob Agents Chemother 54:3967–3969. https://doi.org/10.1128/AAC.00137-10
Zhang D, Yin Z, Zhao Y, Feng J, Jiang X, Zhan Z et al (2017) p1220-CTXM, a pKP048-related IncFIIK plasmid carrying bla CTX-M-14 and qnrB4. Future Microbiol 12:1035–1043. https://doi.org/10.2217/fmb-2017-0026
Vanwynsberghe T, Verhamme K, Raymaekers M, Cartuyvels R, Boel A, De Beenhouwer H (2007) Outbreak of Klebsiella pneumoniae strain harbouring an AmpC (DHA-1) and a bla SHV-11 in a Belgian hospital, August–December 2006. Eur Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull 12:E070201.3
Mata C, Miró E, Toleman M, Rivera MA, Walsh TR, Navarro F (2011) Association of bla(DHA-1) and qnrB genes carried by broad-host-range plasmids among isolates of Enterobacteriaceae at a Spanish hospital. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis 17:1514–1517. https://doi.org/10.1111/j.1469-0691.2011.03539.x
Diestra K, Miró E, Martí C, Navarro D, Cuquet J, Coll P et al (2011) Multiclonal epidemic of Klebsiella pneumoniae isolates producing DHA-1 in a Spanish hospital. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis 17:1032–1036. https://doi.org/10.1111/j.1469-0691.2010.03319.x
Yu F, Chen C, Chen Q, Yu X, Ding B, Yang L et al (2012) Identification of transferable DHA-1 type AmpC β-lactamases and two mutations in quinolone resistance-determining regions of Salmonella enterica serovar Thompson. J Med Microbiol 61:460–462. https://doi.org/10.1099/jmm.0.040691-0
Yim G, Kwong W, Davies J, Miao V (2013) Complex integrons containing qnrB4-ampC (bla(DHA-1)) in plasmids of multidrug-resistant Citrobacter freundii from wastewater. Can J Microbiol 59:110–116. https://doi.org/10.1139/cjm-2012-0576
Pérez-Moreno MO, Estepa V, Sáenz Y, Cortell-Ortolá M, Fort-Gallifa I, Ruiz J et al (2012) Intrahospitalary dissemination of Klebsiella pneumoniae carrying bla(DHA-1) and qnrB4 genes within a novel complex class 1 integron. Diagn Microbiol Infect Dis 73:210–211. https://doi.org/10.1016/j.diagmicrobio.2012.02.011
Guo Q, Wang P, Ma Y, Yang Y, Ye X, Wang M (2012) Co-production of SFO-1 and DHA-1 β-lactamases and 16S rRNA methylase ArmA in clinical isolates of Klebsiella pneumoniae. J Antimicrob Chemother 67:2361–2366. https://doi.org/10.1093/jac/dks244
Compain F, Frangeul L, Drieux L, Verdet C, Brisse S, Arlet G et al (2014) Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrob Agents Chemother 58:4207–4210. https://doi.org/10.1128/AAC.02773-13
Tobes R, Codoñer FM, López-Camacho E, Salanueva IJ, Manrique M, Brozynska M et al (2013) Genome sequence of Klebsiella pneumoniae KpQ3, a DHA-1 β-lactamase-producing nosocomial isolate. Genome Announc 1. https://doi.org/10.1128/genomeA.00167-12
Compain F, Decré D, Fulgencio J-P, Berraho S, Arlet G, Verdet C (2014) Molecular characterization of DHA-1-producing Klebsiella pneumoniae isolates collected during a 4-year period in an intensive care unit. Diagn Microbiol Infect Dis 80:159–161. https://doi.org/10.1016/j.diagmicrobio.2014.06.009
Hidalgo L, Gutierrez B, Ovejero CM, Carrilero L, Matrat S, Saba CKS et al (2013) Klebsiella pneumoniae sequence type 11 from companion animals bearing ArmA methyltransferase, DHA-1 β-lactamase, and QnrB4. Antimicrob Agents Chemother 57:4532–4534. https://doi.org/10.1128/AAC.00491-13
Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I et al (2011) Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989. https://doi.org/10.1371/journal.pone.0017989
Voulgari E, Poulou A, Dimitroulia E, Politi L, Ranellou K, Gennimata V et al (2015) Emergence of OXA-162 carbapenemase- and DHA-1 Amp. cephalosporinase-producing sequence type 11 Klebsiella pneumoniae causing community-onset infection in Greece. Antimicrob Agents Chemother 60:1862–1864. https://doi.org/10.1128/AAC.01514-15
Kuai S, Shao H, Huang L, Pei H, Lu Z, Wang W et al (2014) KPC-2 carbapenemase and DHA-1 Amp. determinants carried on the same plasmid in Enterobacter aerogenes. J Med Microbiol 63:367–370. https://doi.org/10.1099/jmm.0.054627-0
Schlüter A, Nordmann P, Bonnin RA, Millemann Y, Eikmeyer FG, Wibberg D et al (2014) IncH-type plasmid harboring bla CTX-M-15 , bla DHA-1 , and qnrB4 genes recovered from animal isolates. Antimicrob Agents Chemother 58:3768–3773. https://doi.org/10.1128/AAC.02695-14
Kis Z, Tóth Á, Jánvári L, Damjanova I (2016) Countrywide dissemination of a DHA-1-type plasmid-mediated AmpC β-lactamase-producing Klebsiella pneumoniae ST11 international high-risk clone in Hungary, 2009-2013. J Med Microbiol 65:1020–1027. https://doi.org/10.1099/jmm.0.000302
Taniguchi Y, Maeyama Y, Ohsaki Y, Hayashi W, Osaka S, Koide S et al (2017) Co-resistance to colistin and tigecycline by disrupting mgrB and ramR with IS insertions in a canine Klebsiella pneumoniae ST37 isolate producing SHV-12, DHA-1 and FosA3. Int J Antimicrob Agents 50:697–698. https://doi.org/10.1016/j.ijantimicag.2017.09.011
Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L et al (2014) In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. https://doi.org/10.1128/AAC.02412-14
McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ et al (2013) The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. https://doi.org/10.1128/AAC.00419-13
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C et al (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542. https://doi.org/10.1093/nar/gkw1017
Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. https://doi.org/10.1371/journal.pone.0011147
Lawlor MS, O’connor C, Miller VL (2007) Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect Immun 75:1463–1472. https://doi.org/10.1128/IAI.00372-06
Nassif X, Sansonetti PJ (1986) Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect Immun 54:603–608
Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL (2010) RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 192:3144–3158. https://doi.org/10.1128/JB.00031-10
Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J (2009) DNAPlotter: circular and linear interactive genome visualization. Bioinforma Oxf Engl 25(1):119–120. https://doi.org/10.1093/bioinformatics/btn578
Chérif T, Saidani M, Decré D, Boutiba-Ben Boubaker I, Arlet G (2015) Cooccurrence of multiple AmpC β-lactamases in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis in Tunisia. Antimicrob Agents Chemother 60:44–51. https://doi.org/10.1128/AAC.00828-15
Toleman MA, Bennett PM, Walsh TR (2006) ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev MMBR 70:296–316. https://doi.org/10.1128/MMBR.00048-05
Cheng C, Sun J, Zheng F, Lu W, Yang Q, Rui Y (2016) New structures simultaneously harboring class 1 integron and ISCR1-linked resistance genes in multidrug-resistant Gram-negative bacteria. BMC Microbiol 16:71. https://doi.org/10.1186/s12866-016-0683-x
Harmer CJ, Hall RM (2016) IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 1. https://doi.org/10.1128/mSphere.00038-16
Hennequin C, Robin F, Cabrolier N, Bonnet R, Forestier C (2012) Characterization of a DHA-1-producing Klebsiella pneumoniae strain involved in an outbreak and role of the AmpR regulator in virulence. Antimicrob Agents Chemother 56:288–294. https://doi.org/10.1128/AAC.00164-11
Luk S, Wong W-K, Ho AY-M, Yu KC-H, To W-K, Ng T-K (2016) Clinical features and molecular epidemiology of plasmid-mediated DHA-type AmpC β-lactamase-producing Klebsiella pneumoniae blood culture isolates, Hong Kong. J Glob Antimicrob Resist 7:37–42. https://doi.org/10.1016/j.jgar.2016.06.006
Ku Y-H, Chuang Y-C, Chen C-C, Lee M-F, Yang Y-C, Tang H-J et al (2017) Klebsiella pneumoniae isolates from meningitis: epidemiology, virulence and antibiotic resistance. Sci Rep 7:6634. https://doi.org/10.1038/s41598-017-06878-6
Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard A-S et al (2014) Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. https://doi.org/10.3201/eid2011.140206
Cheong HS, Chung DR, Lee C, Kim SH, Kang C-I, Peck KR et al (2016) Emergence of serotype K1 Klebsiella pneumoniae ST23 strains co-producing the plasmid-mediated AmpC beta-lactamase DHA-1 and an extended-spectrum beta-lactamase in Korea. Antimicrob Resist Infect Control 5:50. https://doi.org/10.1186/s13756-016-0151-2
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(ODS 7 kb)
Rights and permissions
About this article
Cite this article
Hennequin, C., Ravet, V. & Robin, F. Plasmids carrying DHA-1 β-lactamases. Eur J Clin Microbiol Infect Dis 37, 1197–1209 (2018). https://doi.org/10.1007/s10096-018-3231-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-018-3231-9