Abstract
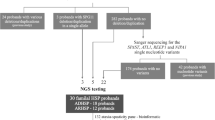
Hereditary spastic paraplegias (HSP) are clinical and genetic heterogeneous diseases with more than 80 disease genes identified thus far. Studies on large cohorts of HSP patients showed that, by means of current technologies, the percentage of genetically solved cases is close to 50%. Notably, the percentage of molecularly confirmed diagnoses decreases significantly in sporadic patients. To describe our diagnostic molecular genetic approach on patients with pediatric-onset pure and complex HSP, 47 subjects with HSP underwent molecular screening of 113 known and candidate disease genes by targeted capture and massively parallel sequencing. Negative cases were successively analyzed by multiplex ligation-dependent probe amplification (MLPA) analysis for the SPAST gene and high-resolution SNP array analysis for genome-wide CNV detection. Diagnosis was molecularly confirmed in 29 out of 47 (62%) patients, most of whom had clinical diagnosis of cHSP. Although SPG11 and SPG4 remain the most frequent cause of, respectively, complex and pure HSP, a large number of pathogenic variants were disclosed in POLR3A, FA2H, DDHD2, ATP2B4, ENTPD1, ERLIN2, CAPN1, ALS2, ADAR1, RNASEH2B, TUBB4A, ATL1, and KIF1A. In a subset of these disease genes, phenotypic expansion and novel genotype-phenotype correlations were recognized. Notably, SNP array analysis did not provide any significant contribution in increasing the diagnostic yield. Our findings document the high diagnostic yield of targeted sequencing for patients with pediatric-onset, complex, and pure HSP. MLPA for SPAST and SNP array should be limited to properly selected cases based on clinical suspicion.


Similar content being viewed by others
References
Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A (2014) Hereditaryspastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms. Exp Neurol 261:518–539
Schüle R, Wiethoff S, Martus P, Karle KN, Otto S, Klebe S, Klimpe S, Gallenmüller C, Kurzwelly D, Henkel D, Rimmele F, Stolze H, Kohl Z, Kassubek J, Klockgether T, Vielhaber S, Kamm C, Klopstock T, Bauer P, Züchner S, Liepelt-Scarfone I, Schöls L (2016) Hereditary spastic paraplegia: clinico-genetic lessons from 608 patients. Ann Neurol 79:646–658
Kara E, Tucci A, Manzoni C, Lynch DS, Elpidorou M, Bettencourt C, Chelban V, Manole A, Hamed SA, Haridy NA, Federoff M, Preza E, Hughes D, Pittman A, Jaunmuktane Z, Brandner S, Xiromerisiou G, Wiethoff S, Schottlaender L, Proukakis C, Morris H, Warner T, Bhatia KP, Korlipara LV, Singleton AB, Hardy J, Wood NW, Lewis PA, Houlden H (2016) Genetic and phenotypic characterization of complex hereditary spastic paraplegia. Brain 139:1904–1918
Povysil G, Tzika A, Vogt J, Haunschmid V, Messiaen L, Zschocke J, Klambauer G, Hochreiter S, Wimmer K (2017) Panelcn.MOPS: copy-number detection in targeted NGS panel data for clinical diagnostics. Hum Mutat 38:889–897
Travaglini L, Bellacchio E, Aiello C, Pro S, Bertini E, Nicita F (2017) Expanding the clinical phenotype of CAPN1-associated mutations: a new case with congenital-onset pure spastic paraplegia. J Neurol Sci 378:210–212
Crow YJ, Zaki MS, Abdel-Hamid MS et al (2014) Mutations in ADAR1, IFIH1, and RNASEH2B presenting as spastic paraplegia. Neuropediatrics 45:386–391
Minnerop M, Kurzwelly D, Wagner H, Soehn AS, Reichbauer J, Tao F, Rattay TW, Peitz M, Rehbach K, Giorgetti A, Pyle A, Thiele H, Altmüller J, Timmann D, Karaca I, Lennarz M, Baets J, Hengel H, Synofzik M, Atasu B, Feely S, Kennerson M, Stendel C, Lindig T, Gonzalez MA, Stirnberg R, Sturm M, Roeske S, Jung J, Bauer P, Lohmann E, Herms S, Heilmann-Heimbach S, Nicholson G, Mahanjah M, Sharkia R, Carloni P, Brüstle O, Klopstock T, Mathews KD, Shy ME, de Jonghe P, Chinnery PF, Horvath R, Kohlhase J, Schmitt I, Wolf M, Greschus S, Amunts K, Maier W, Schöls L, Nürnberg P, Zuchner S, Klockgether T, Ramirez A, Schüle R (2017) Hypomorphic mutations in POLR3A are a frequent cause of sporadic and recessive spastic ataxia. Brain 140:1561–1578
Wakil SM, Bohlega S, Hagos S, Baz B, al Dossari H, Ramzan K, al-Hassnan ZN (2013) A novel splice site mutation in ERLIN2 causes hereditary spastic paraplegia in a Saudi family. Eur J Med Genet 56:43–45
Hehr U, Bauer P, Winner B, Schule R, Olmez A, Koehler W, Uyanik G, Engel A, Lenz D, Seibel A, Hehr A, Ploetz S, Gamez J, Rolfs A, Weis J, Ringer TM, Bonin M, Schuierer G, Marienhagen J, Bogdahn U, Weber BH, Topaloglu H, Schols L, Riess O, Winkler J (2007) Long-term course and mutational spectrum of spatacsin-linked spastic paraplegia. Ann Neurol 62:656–665
Stromillo ML, Malandrini A, Dotti MT, Battaglini M, Borgogni F, Tessa A, Storti E, Denora PS, Santorelli FM, Gaudiano C, Battisti C, Federico A, de Stefano N (2011) Structural and metabolic damage in brains of patients with SPG11-related spastic paraplegia as detected by quantitative MRI. J Neurol 258:2240–2247
Guidubaldi A, Piano C, Santorelli FM, Silvestri G, Petracca M, Tessa A, Bentivoglio AR (2011) Novel mutations in SPG11 cause hereditary spastic paraplegia associated with early-onset levodopa-responsive parkinsonism. Mov Disord 26:553–556
Lee MJ, Cheng TW, Hua MS et al (2008) Mutations of the SPG11 gene in patients with autosomal recessive spastic paraparesis and thin corpus callosum. J Neurol Neurosurg Psychiatry 79:607–609
Denora PS, Schlesinger D, Casali C, Kok F, Tessa A, Boukhris A, Azzedine H, Dotti MT, Bruno C, Truchetto J, Biancheri R, Fedirko E, di Rocco M, Bueno C, Malandrini A, Battini R, Sickl E, de Leva MF, Boespflug-Tanguy O, Silvestri G, Simonati A, Said E, Ferbert A, Criscuolo C, Heinimann K, Modoni A, Weber P, Palmeri S, Plasilova M, Pauri F, Cassandrini D, Battisti C, Pini A, Tosetti M, Hauser E, Masciullo M, di Fabio R, Piccolo F, Denis E, Cioni G, Massa R, Della Giustina E, Calabrese O, Melone MA, de Michele G, Federico A, Bertini E, Durr A, Brockmann K, van der Knaap M, Zatz M, Filla A, Brice A, Stevanin G, Santorelli FM (2009) Screening of ARHSP-TCC patients expands the spectrum of mutations and includes a large scale gene deletion. Hum Mutat 30:E500–E519
Smith BN, Bevan S, Vance C, Renwick P, Wilkinson P, Proukakis C, Squitieri F, Berardelli A, Warner TT, Reid E, Shaw CE (2009) Four novel SPG3A/atlastin mutations identified in autosomal dominant hereditary spastic paraplegia kindreds with intra-familial variability in age of onset and complex phenotype. Clin Genet 75:485–489
Dürr A, Camuzat A, Colin E, Tallaksen C, Hannequin D, Coutinho P, Fontaine B, Rossi A, Gil R, Rousselle C, Ruberg M, Stevanin G, Brice A (2004) Atlastin1 mutations are frequent in young-onset autosomal dominant spastic paraplegia. Arch Neurol 61:1867–1872
Kancheva D, Chamova T, Guergueltcheva V, Mitev V, Azmanov DN, Kalaydjieva L, Tournev I, Jordanova A (2015) Mosaic dominant TUBB4A mutation in an inbred family with complicated hereditary spastic paraplegia. Mov Disord 30:854–858
Livingston JH, Lin JP, Dale RC, Gill D, Brogan P, Munnich A, Kurian MA, Gonzalez-Martinez V, de Goede CG, Falconer A, Forte G, Jenkinson EM, Kasher PR, Szynkiewicz M, Rice GI, Crow YJ (2014) A type I interferon signature identifies bilateral striatal necrosis due to mutations in ADAR1. J Med Genet 51:76–82
Fogli A, Battini R, Baldinotti F, Angela M, Elena CM, Paolo S (2009) Novel human pathological mutations. Gene symbol: SPG4. Disease: spastic paraplegia, autosomal dominant. Hum Genet 126:341
Wang D, Pascual JM, Yang H, Engelstad K, Jhung S, Sun RP, de Vivo DC (2005) Glut-1 deficiency syndrome: clinical, genetic, and therapeutic aspects. Ann Neurol 57:111–118
Fonknechten N, Mavel D, Byrne P, Davoine CS, Cruaud C, Bönsch D, Samson D, Coutinho P, Hutchinson M, McMonagle P, Burgunder JM, Tartaglione A, Heinzlef O, Feki I, Deufel T, Parfrey N, Brice A, Fontaine B, Prud'homme J, Weissenbach J, Dürr A, Hazan J (2000) Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum Mol Genet 9:637–644
Nicita F, Bertini E, Travaglini L, Armando M, Aiello C (2016) Congenital-onset spastic paraplegia in a patient with TUBB4A mutation and mild hypomyelination. J Neurol Sci 368:145–146
Synofzik M, Schüle R (2017) Overcoming the divide between ataxias and spastic paraplegias: shared phenotypes, genes, and pathways. Mov Disord 32:332–345
Kumar KR, Wali GM, Kamate M, Wali G, Minoche AE, Puttick C, Pinese M, Gayevskiy V, Dinger ME, Roscioli T, Sue CM, Cowley MJ (2016) Defining the genetic basis of early onset hereditary spastic paraplegia using whole genome sequencing. Neurogenetics 17:265–270
Lynch DS, Koutsis G, Tucci A, Panas M, Baklou M, Breza M, Karadima G, Houlden H (2016) Hereditary spastic paraplegia in Greece: characterisation of a previously unexplored population using next-generation sequencing. Eur J Hum Genet 24:857–863
Li M, Ho PW, Pang SY, Tse ZH, Kung MH, Sham PC, Ho SL (2014) PMCA4 (ATP2B4) mutation in familial spastic paraplegia. PLoS One 9:e104790
Ho PW, Pang SY, Li M, Tse ZH, Kung MH, Sham PC, Ho SL (2015) PMCA4 (ATP2B4) mutation in familial spastic paraplegia causes delay in intracellular calcium extrusion. Brain Behav 5:e00321
Alazami AM, Adly N, Al Dhalaan H, Alkuraya FS (2011) A nullimorphic ERLIN2 mutation defines a complicated hereditary spastic paraplegia locus (SPG18). Neurogenetics 12:333–336
Al-Saif A, Bohlega S, Al-Mohanna F (2012) Loss of ERLIN2 function leads to juvenile primary lateral sclerosis. Ann Neurol 72:510–516
Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Déry C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martínez-Frías ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP (2006) Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nature Genet 38:910–916
La Piana R, Cayami FK, Tran LT et al (2016) Diffuse hypomyelination is not obligate for POLR3-related disorders. Neurology 86:1622–1626
Beetz C, Nygren AO, Schickel J, Auer-Grumbach M, Bürk K, Heide G, Kassubek J, Klimpe S, Klopstock T, Kreuz F, Otto S, Schüle R, Schöls L, Sperfeld AD, Witte OW, Deufel T (2006) High frequency of partial SPAST deletions in autosomal dominant hereditary spastic paraplegia. Neurology 67:1926–1930
Sulek A, Elert E, Rajkiewicz M, Zdzienicka E, Stepniak I, Krysa W, Zaremba J (2013) Screening for the hereditary spastic paraplaegias SPG4 and SPG3A with the multiplex ligation-dependent probe amplification technique in a large population of affected individuals. Neurol Sci 34:239–242
Mészárosová AU, Putzová M, Čermáková M, Vávrová D, Doležalová K, Smetanová I, Stejskal D, Beetz C, Seeman P (2016) SPAST mutation spectrum and familial occurrence among Czech patients with pure hereditary spastic paraplegia. J Hum Genet 61:845–850
Battini R, Fogli A, Borghetti D, Michelucci A, Perazza S, Baldinotti F, Conidi ME, Ferreri MI, Simi P, Cioni G (2011) Clinical and genetic findings in a series of Italian children with pure hereditary spastic paraplegia. Eur J Neurol 18:150–157
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Integrity of research and reporting
The authors declare that all human and animal studies have been approved by the appropriate ethics committee and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. All the parents of individuals enrolled in the study gave their informed consent prior to their inclusion in the study. Details that might disclose the identity of the subjects under study were omitted.
Electronic supplementary material
ESM 1
(DOCX 326 kb)
Rights and permissions
About this article
Cite this article
Travaglini, L., Aiello, C., Stregapede, F. et al. The impact of next-generation sequencing on the diagnosis of pediatric-onset hereditary spastic paraplegias: new genotype-phenotype correlations for rare HSP-related genes. Neurogenetics 19, 111–121 (2018). https://doi.org/10.1007/s10048-018-0545-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-018-0545-9