Abstract
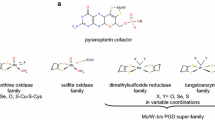
The formate dehydrogenase enzyme from Cupriavidus necator (FdsABG) carries out the two-electron oxidation of formate to CO2, but is also capable of reducing CO2 back to formate, a potential biofuel. FdsABG is a heterotrimeric enzyme that performs this transformation using nine redox-active cofactors: a bis(molybdopterin guanine dinucleotide) (bis-MGD) at the active site coupled to seven iron–sulfur clusters, and one equivalent of flavin mononucleotide (FMN). To better understand the pathway of electron flow in FdsABG, the reduction potentials of the various cofactors were examined through direct electrochemistry. Given the redundancy of cofactors, a truncated form of the FdsA subunit was developed that possesses only the bis-MGD active site and a singular [4Fe–4S] cluster. Electrochemical characterization of FdsABG compared to truncated FdsA shows that the measured reduction potentials are remarkably similar despite the truncation with two observable features at − 265 mV and − 455 mV vs SHE, indicating that the voltammetry of the truncated enzyme is representative of the reduction potentials of the intact heterotrimer. By producing truncated FdsA without the necessary maturation factors required for bis-MGD insertion, a form of the truncated FdsA that possesses only the [4Fe–4S] was produced, which gives a single voltammetric feature at − 525 mV, allowing the contributions of the molybdenum cofactor to be associated with the observed feature at − 265 mV. This method allowed for the deconvolution of reduction potentials for an enzyme with highly complex cofactor content to know more about the thermodynamic landscape of catalysis.







Similar content being viewed by others
References
Solomon S, Plattner G-K, Knutti R, Friedlingstein P (2009) Proc Natl Acad Sci USA 106:1704
Müller J, MacEachran D, Burd H, Sathitsuksanoh N, Bi C, Yeh Y-C, Lee TS, Hillson NJ, Chhabra SR, Singer SW, Beller HR (2013) Appl Environ Microbiol 79:4433
Hille R, Hall J, Basu P (2014) Chem Rev 114:3963–4038
Niks D, Duvvuru J, Escalona M, Hille R (2016) J Biol Chem 291:1162–1174
Yu X, Niks D, Mulchandani A, Hille R (2017) J Biol Chem 292:16872–16879
Oh J-I, Bowien B (1998) J Biol Chem 273:26349–26360
Hartmann T, Leimkühler S (2013) FEBS J 280:6083–6096
Thomé R, Gust A, Toci R, Mendel R, Bittner F, Magalon A, Walburger A (2012) J Biol Chem 287:4671–4678
Akhtar MK, Jones PR (2008) Appl Microbiol Biotechnol 78:853–862
Friedebold J, Mayer F, Bill E, Trautwein AX, Bowien B (1995) Biol Chem Hoppe-Seyler 376:561–568
Carter P (1971) Anal Biochem 40:450–458
McDowall JS, Murphy BJ, Haumann M, Palmer T, Armstrong FA, Sargent F (2014) Proc Natl Acad Sci 111:E3948–E3956
Ayikpoe R, Ngendahimana T, Langton M, Bonitatibus S, Walker LM, Eaton SS, Eaton GR, Pandelia M-E, Elliott SJ, Latham JA (2019) Biochemistry 58:940–950. https://doi.org/10.1021/acs.biochem.8b01082
Walker LM, Kincannon WM, Bandarian V, Elliott SJ (2018) Biochemistry 57:6050–6053
McDowall JS, Murphy BJ, Haumann M, Palmer T, Armstrong FA, Sargent F (2014) Proc Natl Acad Sci USA 111:E3948
Fourmond V (2016) Anal Chem 88:5050–5052
Bassegoda A, Madden C, Wakerley DW, Reisner E, Hirst J (2014) J Am Chem Soc 136:15473–15476
Reda T, Plugge CM, Abram NJ, Hirst J (2008) Proc Natl Acad Sci USA 105:10654
Schlindwein C, Giordano G, Santini CL, Mandrand MA (1990) J Bacteriol 172:6112–6121
Adamson H, Simonov AN, Kierzek M, Rothery RA, Weiner JH, Bond AM, Parkin A (2015) Proc Natl Acad Sci USA 112:14506
McGrath AP, Laming EL, Garcia GPC, Kvansakul M, Guss JM, Trewhella J, Calmes B, Bernhardt PV, Hanson GR, Kappler U, Maher MJ (2015) eLife 4:e09066
Rothery RA, Magalon A, Giordano G, Guigliarelli B, Blasco F, Weiner JH (1998) J Biol Chem 273:7462–7469
Wu S-Y, Rothery RA, Weiner JH (2015) J Biol Chem 290:25164–25173
Heering HA, Hagen WR (1996) J Electroanal Chem 404:249–260
Baymann F, Schoepp-Cothenet B, Duval S, Guiral M, Brugna M, Baffert C, Russell MJ, Nitschke W (2018) Frontiers in Microbiology 9:1357
Lim ZH, Chng ELK, Hui Y, Webster RD (2013) J Phys Chem B 117:2396–2402
Tan SLJ, Novianti ML, Webster RD (2015) J Phys Chem B 119:14053–14064
Acknowledgements
This work was supported by the Department of Energy, Office of Sciences, Basic Energy Sciences (BES) program, via contract BES DE-SC0012598 (to SJE) and DE-SC0010666 (to RH).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Walker, L.M., Li, B., Niks, D. et al. Deconvolution of reduction potentials of formate dehydrogenase from Cupriavidus necator. J Biol Inorg Chem 24, 889–898 (2019). https://doi.org/10.1007/s00775-019-01701-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-019-01701-1