Abstract
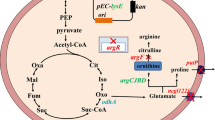
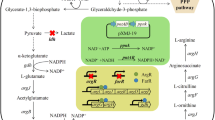
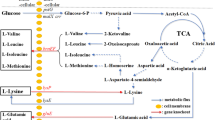
The experiments presented here were based on the conclusions of our previous results. In order to avoid introduction of expression plasmid and to balance the NADH/NAD ratio, the NADH biosynthetic enzyme, i.e., NAD-dependent glyceraldehyde-3-phosphate dehydrogenase (GADPH), was replaced by NADP-dependent GADPH, which was used to biosynthesize NADPH rather than NADH. The results indicated that the NADH/NAD ratio significantly decreased, and glucose consumption and l-lysine production drastically improved. Moreover, increasing the flux through l-lysine biosynthetic pathway and disruption of ilvN and hom, which involve in the branched amino acid and l-methionine biosynthesis, further improved l-lysine production by Corynebacterium glutamicum. Compared to the original strain C. glutamicum Lys5, the l-lysine production and glucose conversion efficiency (α) were enhanced to 81.0 ± 6.59 mM and 36.45 % by the resulting strain C. glutamicum Lys5-8 in shake flask. In addition, the by-products (i.e., l-threonine, l-methionine and l-valine) were significantly decreased as results of genetic modification in homoserine dehydrogenase (HSD) and acetohydroxyacid synthase (AHAS). In fed-batch fermentation, C. glutamicum Lys5-8 began to produce l-lysine at post-exponential growth phase and continuously increased over 36 h to a final titer of 896 ± 33.41 mM. The l-lysine productivity was 2.73 g l−1 h−1 and the α was 47.06 % after 48 h. However, the attenuation of MurE was not beneficial to increase the l-lysine production because of decreasing the cell growth. Based on the above-mentioned results, we get the following conclusions: cofactor NADPH, precursor, the flux through l-lysine biosynthetic pathway and DCW are beneficial to improve l-lysine production in C. glutamicum.




Similar content being viewed by others
References
Becker J, Klopprogge C, Herold A, Zelder O, Bolten CJ, Wittmann C (2007) Metabolic flux engineering of l-lysine production in Corynebacterium glutamicum—over expression and modification of G6P dehydrogenase. J Biotechnol 138(2):99–109. doi:10.1016/j.jbiotec.2007.05.026
Becker J, Zelder O, Häfner S, Schröder H, Wittmann C (2011) From zero to hero-design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metab Eng 13(2):159–168. doi:10.1016/j.ymben.2011.01.003
Binder S, Schendzielorz G, Stäbler N, Krumbach K, Hoffmann K, Bott M, Eggeling L (2012) A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol 13:R40. doi:10.1186/gb-2012-13-5-r40
Blombach B, Schreiner ME, Moch M, Oldiges M, Eikmanns BJ (2007) Effect of pyruvate dehydrogenase complex deficiency on l-lysine production with Corynebacterium glutamicum. Appl Microbiol Biotechnol 76(3):615–623. doi:10.1007/s00253-007-0904-1
Blombach B, Hans S, Bathe B, Eikmanns BJ (2009) Acetohydroxyacid synthase, a novel target for improvement of l-lysine production by Corynebacterium glutamicum. Appl Environ Microbiol 75(2):419–427. doi:10.1128/AEM.01844-08
Chen Z, Rappert S, Zeng AP (2013) Rational design of allosteric regulation of homoserine dehydrogenase by a nonnatural inhibitor l-lysine. ACS Synth Biol. doi:10.1021/sb400133g
Dong X, Quinn PJ, Wang X (2011) Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for the production of l-threonine. Biotechnol Adv 29(1):11–23. doi:10.1016/j.biotechadv.2010.07.009
Fiuza M, Canova MJ, Patin D, Letek M, Zanella-Cléon I, Becchi M, Mateos LM, Mengin-Lecreulx D, Molle V, Gil JA (2008) The MurC ligase essential for peptidoglycan biosynthesis is regulated by the serine/threonine protein kinase PknA in Corynebacterium glutamicum. J Biol Chem 283(52):36553–36563. doi:10.1074/jbc.M807175200
Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, Takio S, Shinozaki K, Takano H (2008) An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J 53(6):924–934. doi:10.1111/j.1365-313X.2007.03379.x
Georgi T, Rittmann D, Wendisch VF (2005) Lysine and glutamate production by Corynebacterium glutamicum on glucose, fructose and sucrose: roles of malic enzyme and fructose-1,6-bisphosphatase. Metab Eng 7(4):291–301. doi:10.1016/j.ymben.2005.05.001
Hasegawa S, Suda M, Uematsu K, Natsuma Y, Hiraga K, Jojima T, Inui M (2013) Engineering of Corynebacterium glutamicum for high-yield l-valine production under oxygen deprivation conditions. Appl Environ Microbiol 79(4):1250–1257. doi:10.1128/AEM.02806-12
Hou XH, Chen XD, Zhang Y, Qian H, Zhang WG (2012) (l)-Valine production with minimization of by-products’ synthesis in Corynebacterium glutamicum and Brevibacterium flavum. Amino Acids 43(6):2301–2311. doi:10.1007/s00726-012-1308-9
Jiang LY, Zhang YY, Li Z, Liu JZ (2013) Metabolic engineering of Corynebacterium glutamicum for increasing the production of l-ornithine by increasing NADPH availability. J Ind Microbiol Biotechnol 40(10):1143–1151. doi:10.1007/s10295-013-1306-2
Kabus A, Georgi T, Wendisch VF, Bott M (2007) Expression of the Escherichia coli pntAB genes encoding a membrane-bound transhydrogenase in Corynebacterium glutamicum improves l-lysine formation. Appl Microbiol Biotechnol 75(1):47–53. doi:10.1007/s00253-006-0804-9
Kelle R, Hermann T, Bathe B (2005) l-lysine production. In: Eggeling L, Bott M (eds) Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, pp 465–488
Lee HC, Kim JS, Jang W, Kim SY (2010) High NADPH/NADP+ ratio improves thymidine production by a metabolically engineered Escherichia coli strain. J Biotechnol 149(1–2):24–32. doi:10.1016/j.jbiotec.2010.06.011
Martínez I, Zhu J, Lin H, Bennett GN, San KY (2008) Replacing Escherichia coli NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with an NADP-dependent enzyme from Clostridium acetobutylicum facilitates NADPH dependent pathways. Metab Eng 10(6):352–359. doi:10.1016/j.ymben.2008.09.001
Morbach S, Sahm H, Eggeling L (1996) l-isoleucine production with Corynebacterium glutamicum: further flux increase and limitation of export. Appl Environ Microbiol 62(12):4345–4351
Nakotte S, Schaffer S, Bohringer M, Durre P (1998) Electroporation of, plasmid isolation from and plasmid conservation in Clostridium acetobutylicum DSM 792. Appl Microbiol Biotechnol 50(5):564–567. doi:10.1007/s002530051335
Ohnishi J, Mitsuhashi S, Hayashi M, Ando S, Yokoi H, Ochiai K, Ikeda M (2002) A novel methodology employing Corynebacterium glutamicum genome information to generate a new l-lysine-producing mutant. Appl Microbiol Biotechnol 58(2):217–223. doi:10.1007/s00253-001-0883-6
Osman K, Evangelopooulos D, Basavannacharya C, Gupta A, McHugh TD, Bhakta S, Gibbons S (2012) An antibacterial from Hypericum acmosepalum inhibits ATP-dependent MurE ligase from Mycobacterium tuberculosis. Int J Antimicrob Aging 39(2):124–129. doi:10.1016/j.ijantimicag.2011.09.018
Peters-Wendisch PG, Kreutzer C, Kalinowski J, Patek M, Sahm H, Eikmanns BJ (1998) Pyruvate carboxylase from Corynebacterium glutamicum: characterization, expression and inactivation of the pyc gene. Microbiol 144(Pt4):915–927. doi:10.1099/00221287-144-4-915
Pisabarro A, Malumbes M, Mateos LM, Oguiza JA, Martín JF (1993) A cluster of three genes (dapA, orf2, and dapB) of Brevibacterium lactofermentum encodes dihydrodipicolinate synthase, dihydrodipicolinate reductase, and a third polypeptide of unknown function. J Bacteriol 175(9):2743–2749
Sahm H, Eggeling L, de Graaf AA (2000) Pathway analysis and metabolic engineering in Corynebacterium glutamicum. Biol Chem 381(9–10):899–910. doi:10.1515/BC.2000.111
Sambrook J, Russel DV (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutumicum. Gene 145(1):69–73. doi:10.1016/0378-1119(94)90324-7
Takeno S, Murata R, Kobagashi R, Mitsuhashi S, Ikeda M (2010) Engineering of Corynebacterium glutamicum with an NADPH-generating glycolytic pathway for l-lysine production. Appl Environ Microbiol 76(10):7154–7160. doi:10.1016/j.ymben.2008.09.001
van der Restá ME, Lange C, Molenaar D (1999) A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl Microbiol Biotechnol 52(4):541–545. doi:10.1007/s002530051557
Wendisch VF (2007) Amino acid biosynthesis ~ pathways, regulation and metabolic engineering. In: Wittmann C, Becker J (eds) The l-lysine story: from metabolic pathways to industrial production. Springer, Berlin Heidelberg, pp 39–70
Xu DQ, Tan YZ, Huan XJ, Hu XQ, Wang XY (2010) Construction of a novel shuttle vector for use in Brevibacterium flavum, an industrial amino acid producer. J Microbiol Methods 80(1):86–92. doi:10.1016/j.mimet.2009.11.003
Xu JZ, Han M, Zhang JL, Guo YF, Qian H, Zhang WG (2013a) Improvement of l-lysine production combines with minimization of by-products synthesis in Corynebacterium glutamicum. J Chem Technol Biotechnol. doi:10.1002/jctb.4278
Xu JZ, Zhang JL, Guo YF, Zai YG, Zhang WG (2013b) Inprovement of cell growth and production of l-lysine by genetically modified C. glutamicum during growth on molasses. J Ind Microbiol Biotechnol 40(12):1423–1432. doi:10.1007/s10295-013-1329-8
Xu JZ, Xia XH, Zhang JL, Guo YF, Qian H, Zhang WG (2014a) A method for gene amplification and simultaneous deletion in Corynebacterium glutamicum genome without any genetic markers. Plasmid 72(1):9–17. doi:10.1016/j.plasmid.2014.02.001
Xu JZ, Zhang JL, Guo YF, Zhang WG (2014b) Genetically modifying aspartate aminotransferase and aspartate ammonia-lyase affects metabolite accumulation in L-lysine producing strain derived from Corynebacterium glutamicum ATCC13032 (unpblished data)
Zhang WW, Jiang WH, Zhao GP, Yang YL, Chiao JS (1999) Sequence analysis and expression of the aspartokinase and aspartate semialdehyde dehydrogenase operon from rifamycin SV-producing Amycolatopsis mediterranei. Gene 237(2):413–419. doi:10.1016/S0378-1119(99)00307-8
Acknowledgments
This work was financially supported by the Program of Chinese 863 National High-Tech Research and Development Plan Project (No. 2008AA02Z212) and by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, J., Han, M., Zhang, J. et al. Metabolic engineering Corynebacterium glutamicum for the l-lysine production by increasing the flux into l-lysine biosynthetic pathway. Amino Acids 46, 2165–2175 (2014). https://doi.org/10.1007/s00726-014-1768-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-014-1768-1