Abstract
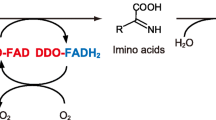
d-Aspartate oxidase (DDO) and d-amino acid oxidase (DAO) are flavin adenine dinucleotide-containing flavoproteins that catalyze the oxidative deamination of d-amino acids. Unlike DAO, which acts on several neutral and basic d-amino acids, DDO is highly specific for acidic d-amino acids. Based on molecular modeling and simulated annealing docking analyses, a recombinant mouse DDO carrying two substitutions (Arg-216 to Leu and Arg-237 to Tyr) was generated (R216L-R237Y variant). This variant and two previously constructed single-point mutants of mouse DDO (R216L and R237Y variants) were characterized to investigate the role of Arg-216 and Arg-237 in the substrate specificity of mouse DDO. The R216L-R237Y and R216L variants acquired a broad specificity for several neutral and basic d-amino acids, and showed a considerable decrease in activity against acidic d-amino acids. The R237Y variant, however, did not show any additional specificity for neutral or basic d-amino acids and its activity against acidic d-amino acids was greatly reduced. The kinetic properties of these variants indicated that the Arg-216 residue is important for the catalytic activity and substrate specificity of mouse DDO. However, Arg-237 is, apparently, only marginally involved in substrate recognition, but is important for catalytic activity. Notably, the substrate specificity of the R216L-R237Y variant differed significantly from that of the R216L variant, suggesting that Arg-237 has subsidiary effects on substrate specificity. Additional experiments using several DDO and DAO inhibitors also suggested the involvement of Arg-216 in the substrate specificity and catalytic activity of mouse DDO and that Arg-237 is possibly involved in substrate recognition by this enzyme. Collectively, these results indicate that Arg-216 and Arg-237 play crucial and subsidiary role(s), respectively, in the substrate specificity of mouse DDO.



Similar content being viewed by others
Abbreviations
- DAO:
-
d-Amino acid oxidase
- DDO:
-
d-Aspartate oxidase
- FAD:
-
Flavin adenine dinucleotide
- IC50 :
-
50% Inhibitory concentration
- IPTG:
-
Isopropyl-β-d-thiogalactopyranoside
- NMDA:
-
N-Methyl-d-aspartate
References
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201
D’Aniello A (2007) d-Aspartic acid: an endogenous amino acid with an important neuroendocrine role. Brain Res Rev 53:215–234
D’Aniello G, Ronsini S, Guida F, Spinelli P, D’Aniello A (2005) Occurrence of d-aspartic acid in human seminal plasma and spermatozoa: possible role in reproduction. Fertil Steril 84:1444–1449
D’Aniello G, Grieco N, Di Filippo MA, Cappiello F, Topo E, D’Aniello E, Ronsini S (2007) Reproductive implication of d-aspartic acid in human pre-ovulatory follicular fluid. Hum Reprod 22:3178–3183
De Marco C, Crifò C (1967) d-Aspartate oxidase from pig kidney. III. Competitive inhibition by dicarboxylic hydroxyacids. Enzymologia 33:325–330
Dixon M, Kenworthy P (1967) d-Aspartate oxidase of kidney. Biochim Biophys Acta 146:54–76
Fonda ML, Anderson BM (1968) d-Amino acid oxidase. II. Studies of substrate-competitive inhibitors. J Biol Chem 243:1931–1935
Homma H (2007) Biochemistry of d-aspartate in mammalian cells. Amino Acids 32:3–11
Huey R, Goodsell DS, Morris GM, Olson AJ (2004) Grid-based hydrogen bond potentials with improved directionality. Lett Drug Des Discov 1:178–183
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28:1145–1152
Katane M, Homma H (2010) d-Aspartate oxidase: the sole catabolic enzyme acting on free d-aspartate in mammals. Chem Biodivers. doi:10.1002/cbdv.200900250
Katane M, Furuchi T, Sekine M, Homma H (2007a) Molecular cloning of a cDNA encoding mouse d-aspartate oxidase and functional characterization of its recombinant proteins by site-directed mutagenesis. Amino Acids 32:69–78
Katane M, Seida Y, Sekine M, Furuchi T, Homma H (2007b) Caenorhabditis elegans has two genes encoding functional d-aspartate oxidases. FEBS J 274:137–149
Katane M, Hanai T, Furuchi T, Sekine M, Homma H (2008) Hyperactive mutants of mouse d-aspartate oxidase: mutagenesis of the active site residue serine 308. Amino Acids 35:75–82
Katane M, Saitoh Y, Seida Y, Sekine M, Furuchi T, Homma H (2010) Comparative characterization of three d-aspartate oxidases and one d-amino acid oxidase from Caenorhabditis elegans. Chem Biodivers. doi:10.1002/cbdv.200900294
Kawazoe T, Tsuge H, Pilone MS, Fukui K (2006) Crystal structure of human d-amino acid oxidase: context-dependent variability of the backbone conformation of the VAAGL hydrophobic stretch located at the si-face of the flavin ring. Protein Sci 15:2708–2717
Kawazoe T, Tsuge H, Imagawa T, Aki K, Kuramitsu S, Fukui K (2007) Structural basis of d-DOPA oxidation by d-amino acid oxidase: alternative pathway for dopamine biosynthesis. Biochem Biophys Res Commun 355:385–391
Klein JR (1960) Competitive inhibition of d-amino acid oxidase by benzoate as a function of substrate. Biochim Biophys Acta 37:534–537
Matsui T, Sekiguchi M, Hashimoto A, Tomita U, Nishikawa T, Wada K (1995) Functional comparison of d-serine and glycine in rodents: the effect on cloned NMDA receptors and the extracellular concentration. J Neurochem 65:454–458
Mattevi A, Vanoni MA, Todone F, Rizzi M, Teplyakov A, Coda A, Bolognesi M, Curti B (1996) Crystal structure of d-amino acid oxidase: a case of active site mirror-image convergent evolution with flavocytochrome b 2. Proc Natl Acad Sci USA 93:7496–7501
Miura R, Setoyama C, Nishina Y, Shiga K, Mizutani H, Miyahara I, Hirotsu K (1997) Structural and mechanistic studies on d-amino acid oxidase-substrate complex: implications of the crystal structure of enzyme-substrate analog complex. J Biochem 122:825–833
Mizutani H, Miyahara I, Hirotsu K, Nishina Y, Shiga K, Setoyama C, Miura R (1996) Three-dimensional structure of porcine kidney d-amino acid oxidase at 3.0 Å resolution. J Biochem 120:14–17
Mothet JP, Parent AT, Wolosker H, Brady RO Jr, Linden DJ, Ferris CD, Rogawski MA, Snyder SH (2000) d-Serine is an endogenous ligand for the glycine site of the N-methyl-d-aspartate receptor. Proc Natl Acad Sci USA 97:4926–4931
Nishikawa T (2005) Metabolism and functional roles of endogenous d-serine in mammalian brains. Biol Pharm Bull 28:1561–1565
Peitsch MC (1995) Protein modeling by E-mail. Biotechnology (N Y) 13:658–660
Pilone MS (2000) d-Amino acid oxidase: new findings. Cell Mol Life Sci 57:1732–1747
Pollegioni L, Piubelli L, Sacchi S, Pilone MS, Molla G (2007) Physiological functions of d-amino acid oxidases: from yeast to humans. Cell Mol Life Sci 64:1373–1394
Sacchi S, Lorenzi S, Molla G, Pilone MS, Rossetti C, Pollegioni L (2002) Engineering the substrate specificity of d-amino-acid oxidase. J Biol Chem 277:27510–27516
Scolari MJ, Acosta GB (2007) d-Serine: a new word in the glutamatergic neuro-glial language. Amino Acids 33:563–574
Setoyama C, Nishina Y, Mizutani H, Miyahara I, Hirotsu K, Kamiya N, Shiga K, Miura R (2006) Engineering the substrate specificity of porcine kidney d-amino acid oxidase by mutagenesis of the “active-site lid”. J Biochem 139:873–879
Tishkov VI, Khoronenkova SV (2005) d-Amino acid oxidase: structure, catalytic mechanism, and practical application. Biochemistry (Mosc) 70:40–54
Todone F, Vanoni MA, Mozzarelli A, Bolognesi M, Coda A, Curti B, Mattevi A (1997) Active site plasticity in d-amino acid oxidase: a crystallographic analysis. Biochemistry 36:5853–5860
Wilkinson GN (1961) Statistical estimations in enzyme kinetics. Biochem J 80:324–332
Acknowledgments
The authors would like to thank Prof. Ryuichi Konno (International University of Health and Welfare) for his kind gift of the mouse DAO expression plasmid, and Mr. Yutaka Taniguchi and Ms. Mayumi Tetsuka, Midori Nitta, and Suno Masakawa for their technical assistance. This work was supported in part by a Grant-in-Aid for Scientific Research (21590071) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, as well as by a Kitasato University Research Grant for Young Researchers (to M.K.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Katane, M., Saitoh, Y., Maeda, K. et al. Role of the active site residues arginine-216 and arginine-237 in the substrate specificity of mammalian d-aspartate oxidase. Amino Acids 40, 467–476 (2011). https://doi.org/10.1007/s00726-010-0658-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-010-0658-4