Abstract
Alanine aminotransferase (AlaAT) catalyses the reversible conversion of pyruvate and glutamate into alanine and oxoglutarate. In soybean, two subclasses were identified, each represented by two highly similar members. To investigate the role of AlaAT during hypoxic stress in soybean, changes in transcript level of both subclasses were analysed together with the enzyme activity and alanine content of the tissue. Moreover, the dependency of AlaAT activity and gene expression was investigated in relation to the source of nitrogen supplied to the plants. Using semi-quantitative PCR, GmAlaAT genes were determined to be highest expressed in roots and nodules. Under normal growth conditions, enzyme activity of AlaAT was detected in all organs tested, with lowest activity in the roots. Upon waterlogging-induced hypoxia, AlaAT activity increased strongly. Concomitantly, alanine accumulated. During re-oxygenation, AlaAT activity remained high, but the transcript level and the alanine content decreased. Our results show a role for AlaAT in the catabolism of alanine during the initial period of re-oxygenation following hypoxia. GmAlaAT also responded to nitrogen availability in the solution during waterlogging. Ammonium as nitrogen source induced both gene expression and enzyme activity of AlaAT more than when nitrate was supplied in the nutrient solution. The work presented here indicates that AlaAT might not only be important during hypoxia, but also during the recovery phase after waterlogging, when oxygen is available to the tissue again.
Similar content being viewed by others
Introduction
Alanine aminotransferase (AlaAT) is a pyridoxal phosphate-dependent enzyme usually found in all plant parts. Its activity is found not only in leaves and roots, but also in other tissues like the endosperm (Kikuchi et al. 1999) and flowers (Igarashi et al. 2003). This broad expression profile of AlaAT indicates that the enzyme is involved in an essential biochemical reaction during the whole life cycle of the plant. Indeed, the enzyme catalyses the reversible reaction between pyruvate and glutamate into alanine and oxoglutarate (EC 2.6.1.2), thereby linking primary carbon metabolism with the synthesis of various amino acids.
The AlaAT is suggested to play a special role during hypoxic stress like it is induced by waterlogging or flooding. It was shown for various plant species that the activity of AlaAT, as well as the accumulation of alanine, increases when the oxygen availability to plant tissues decreases (Good and Muench 1993; Muench and Good 1994; Miyashita et al. 2007; Limami et al. 2008; Rocha et al. 2010). Other metabolic changes that occur simultaneously during hypoxia are a down-regulation of respiratory activity and a decrease of the adenylate energy charge (Gupta et al. 2009; van Dongen et al. 2009), as well as acidification of the cytosol due to lactate accumulation and the loss of carbon due to fermentative production of ethanol (Zabalza et al. 2009). Different to the production of lactate and ethanol, alanine accumulation does not have detrimental side effects to the cell, but rather maintains the glycolytic flux, while retaining carbon and nitrogen resources within the cell (Rocha et al. 2010).
The role of AlaAT during the recovery phase after hypoxic stress is probably equally important for the understanding of flooding tolerance (Fan et al. 1988) as its biochemical behaviour during hypoxia. de Sousa and Sodek (2003) demonstrated that AlaAT activity increased during hypoxia more so after the accumulation of alanine had reached its maximum. Since the level of alanine returned to pre-hypoxic levels within 24 h of return to normoxia, it was suggested that AlaAT has a function during the recovery phase also. Miyashita et al. (2007) confirmed this by showing that the Arabidopsis AlaAT1 knock-out mutant (alaat1-1) was able to accumulate alanine during hypoxia like wild type plants, whereas the decrease of the levels of alanine during the re-oxygenation phase was delayed. It was suggested that during the hypoxic phase, the plant prepares itself for a rapid recovery once oxygen becomes available again. This hypothesis is mainly based on the observation that many genes that encode proteins with an important function during the recovery phase are already expressed during the hypoxic stress (Drew 1997). Furthermore, most metabolites recover rapidly to their normal level once hypoxia is over (Barret-Lennard et al. 1988; Fan et al. 1988; Albrecht et al. 1993; de Sousa and Sodek 2003; Rocha et al. 2010). AlaAT has exactly this kind of regulation pattern as its gene expression is up-regulated during hypoxia, and during the re-oxygenation phase, the high levels of AlaAT enzyme can ensure the rapid conversion of accumulated alanine back into glutamate (Miyashita et al. 2007).
Plants possess a wide variety of aminotransferases that share considerable sequence similarity. Functional analysis of four aminotransferases from Arabidopsis revealed at least two sequences encoding for true AlaAT (E.C.2.6.1.2) enzymes, whereas two homologues that cluster within the same gene-subfamily as the AlaAT genes were shown to act as glutamate:glyoxylate aminotransferase (E.C.2.6.1.4; Igarashi et al. 2003). Subcellular fractionation analysis has shown that the activity of these latter enzymes was principally located in peroxisomes, and it was suggested that these enzymes play an important role in photorespiration and amino acid metabolism (Igarashi et al. 2003; Liepman and Olsen 2003). The other two genes, AtAlaAT1 and AtAlaAT2, were not functionally characterised in detail for Arabidopsis, but in silico prediction of their localisation suggested that AtAlaAT2 is a mitochondrial enzyme and AtAlaAT1 is located in the cytosol (Liepman and Olsen 2003).
Due to its important role in nitrogen and carbon metabolism in plants, AlaAT has been intensively studied in several plant species (de Sousa and Sodek 2003; Ricoult et al. 2006; Good et al. 2007; Miyashita et al. 2007; Beatty et al. 2009). Here, we focus especially on the regulation of AlaATs during hypoxia in relation to the nitrogen status of the plant. We used soybean as symbiotic nitrogen-fixing plants are an ideal model system to investigate the role of nitrate or ammonium as primary nitrogen source. Therefore, we set out to characterise the AlaAT multigene family in soybean plants, and investigated changes in gene expression, enzyme activity and alanine accumulation in various plant organs and under different conditions such as various nitrogen sources and changing oxygen availability.
Materials and methods
Plant material and growth conditions
Soybean plants (Glycine max L. Merril cv. IAC-17) were grown in the greenhouse under natural light and temperature conditions. Three plants were grown together in one plastic pot with a volume of 3 L containing vermiculite as substrate and supplied with 200 mL N-free nutrient solution twice per week (CaCl2 0.5 mM; KCl 0.5 mM; KH2PO4 0.25 mM; K2HPO4 0.25 mM; MgSO4 1.0 mM; FeEDTA 0.05 mM; trace elements: MnCl2 9.1 μM; H3BO3 0.046 mM; ZnCl2 0.765 μM; NaMoO4 0.56 μM; CuCl2 0.32 μM as described by Hoagland and Arnon 1950). For inoculated Bradyrhizobium elkanii strain SEMIA 5019 was used. Shortly before flowering, the pots were transferred into containers, and the root system was flooded with N-free nutrient solution at one-third strength. When the effect of different nitrogen sources for either nodulated as well as non-nodulated plants was tested, KNO3 or (NH4)2SO4 was added to the N-free nutrient solution to a final concentration of 5 mM of nitrogen (as indicated in the text or figures). Waterlogging was maintained during 3 days. Non-waterlogged control plants were set up simultaneously, and supplied with 200 mL full strength N-free nutrient solution as described above. All plants that were cultivated without Bradyrhizobium inoculation were supplied with a nutrient solution supplemented with 15 mM KNO3. At harvest, samples of pods, leaf, root and nodules were taken from 8-week-old plants at the same developmental stage, and frozen in liquid nitrogen and subsequently lyophilised. The lyophilised material was stored at −20°C in a desiccator containing silica gel.
Identification of AlaAT sequences and phylogenetic analysis of AlaAT
The current chromosome-scale assembly (Glyma1.0) of the Soybean genome sequencing project (Schmutz et al. 2010) was used for blasting with the Phytozome v5.0 software (http://www.phytozome.net) against AlaAT cDNA sequences previously identified in Arabidopsis thaliana (Miyashita et al. 2007) and Medicago truncatula (Ricoult et al. 2006).
Phylogenetic analysis of alanine aminotransferase proteins from both mono- and dicotyledonous plant species was performed using amino acid sequences found in the public databases (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi). An un-rooted tree was created applying the neighbour-joining method with MEGA4 (Tamura et al. 2007; http://www.ebi.ac.uk/Tools/clustalw2/index.html). The following protein sequences were included: Glycine max GmAlaAT1 (ABW17196), GmAlaAT2 (ABW17197) and GmAlaAT3 (ABW17198) and the translated amino acid sequence from Glyma16g01630; Arabidopsis thaliana AtAlaAT1 (AAF82782), AtAlaAT2 (NP565040), AtGGT1 (NP564192) and AtGGT2 (NP177215); Medicago truncatula MtmAlaAT and MtcAlaAT (Ricoult et al. 2006); Populus trichocarpa PtAlaAT1 (XP002315675), PtAlaAT2 (XP002312679), PtAlaAT3 (XP002331223) and PtAlaAT4 (XP002304255); Chlamydomonas reinhardtii CrAlaAT1 (XP001695350) and CrAlaAT2 (XP001698518); Oryza sativa cv Japonica Os07g42600 (EEE67593), Os10g25130 (NP001064504), Os03g08530 (ABF94336), Os10g25140 (ABB47495), Os09g26380 (NP001063248) and Os07g01760 (NP001058716); Physcomitrella patens subsp. patens PpAlaAT1 (XP001769989), PpAlaAT2 (XP001753102), PpGGT1 (XP001777071), and PpGGT2 (XP001782822). All sequences were aligned using the programme CLUSTAL (Higgins et al. 1994).
Free amino acid analysis
To determine changes in the alanine content of plants exposed to the various treatments, total free amino acids were extracted with 10 ml of methanol:chloroform:water (12/5/3 v/v) per gram plant material (Bieleski and Turner 1966). The aqueous phase was recovered after phase separation, and individual amino acids were analysed as their OPA derivatives by reverse-phase HPLC, as described previously (Puiatti and Sodek 1999).
Aminotransferase activity assay
The AlaAT enzyme activity (EC 2.6.1.2) was determined in various tissues of soybean as indicated in the text. Plant material was ground with a mortar and pestle in five volumes of 50 mM Tris/HCl pH 7.5 containing 1 mM DDT. All experiments were carried out at 4°C. The homogenate was centrifuged at 10,000g for 20 min, and an aliquot of the supernatant was desalted using a PD10 column (GE Healthcare, Buckinghamshire, UK). Total protein content of the enzyme extract was measured as described by Bradford (1976).
The eluted protein fraction was specifically assayed for AlaAT activity (EC 2.6.1.2) essentially as described by Good and Muench (1992). The AlaAT activity assay contained, in a final volume of 3 ml, 10 mM l-alanine, 5 mM oxoglutarate, 0.1 mM NADH, 50 mM Tris–HCl (pH 7.5) and five units of lactate dehydrogenase type V–S from rabbit muscle (Sigma). After adding extract to the reaction buffer, light absorbtion was determined in 30-s time intervals at 340 nm using a spectrophotometer Ultrospec 1000 (Pharmacia-Biotec, Cambridge, UK) with a temperature-controlled cuvette holder at 30°C.
Semi-quantitative RT-PCR
In order to determine changes of AlaAT gene expression, plant material of soybean was ground in liquid nitrogen using a pestle and mortar. For RNA extraction 0.9 g of root or 0.6 g of nodule, leaf or pod was mixed with 3 mL of TRIzol Reagent (Invitrogen, Carlsbad, USA) following the manufacturer’s recommendations. The RNA samples were stored at −80°C until further processing. For the synthesis of cDNA, 5.0 μg of total RNA was first treated with Turbo DNA-free (Ambion, Austin, USA) to remove DNA contamination. Subsequently, cDNA was synthesised using oligo(dt) 12–18 primers and Superscript II reverse transcriptase (Invitrogen, Carlsbad, USA). Subsequently, the cDNA was used as template for a polymerase chain reaction (PCR) in a total volume of 25 μl with the following reaction conditions: an initial step of 3 min at 96°C was followed by a repeating cycle of 1 min at 94°C, 1 min at 60°C and 1 min at 72°C. Primer selection was performed using the programme Lasergene (DNASTAR, Madison, USA) against GmAlaAT1 (EU165371) and GmAlaAT2 EU165372cDNA sequences from soybean which are representative for the gene subfamilies A and B, respectively. The primer sequences were as follows: GmAlaAT1: sense: CTTCTGCGCCACCGTCACCA antisense: CAGGGCTTGCACCATCAGTCAT; GmAlaAT2: sense: TGGCCACAATTGAAGGACGAG, antisense: GCATCAGCAGGGAATAGCAGT; Actin: sense: GCTCCTAGGGCTGTCTTTCC, antisense: CTCAGCAGAGGTGGTGAACA. Preliminary experiments were performed to identify the optimal cycle number at which non-saturated signal intensities were obtained for all ethidium bromide stained PCR products after electrophoresis in a 1% agarose gel TAE buffer. The number of cycles that was finally chosen is indicated next to the gel figures shown in the results section. Actin was used as a loading control to be used for normalisation of the data.
Results
In silico identification and characterisation of AlaAT genes in soybean
From the soybean (G. max cv. Williams 82) genome sequencing information available in the public data domain, it was searched for the presence of AlaAT homologs using the BLAST tool. Four sequences were found (Glyma01g03260, Glyma02g04320, Glyma07g05130 and Glyma16g01630), each of them located at a different chromosome. For three of these genomic sequences, full length coding sequences were already annotated from EST contigs in the NCBI database (National Centre for Biotechnology Information, http://www.ncbi.nlm.nih.gov) with the following names: Glyma07g05130, GmAlaAT1 (ABW17196); Glyma02g04320, GmAlaAT2 (ABW17197) and Glyma01g03260, GmAlaAT3 (ABW17198). The fourth genomic sequence we identified (Glyma16g01630) was annotated here as GmAlaAT4.
The Glyma02g04320 (GmAlaAT2) and Glyma01g03260 (GmAlaAT3) formed a pair of highly similar sequences both in coding as well as non-coding regions, and so did Glyma07g05130 (GmAlaAT1) and Glyma16g01630 (GmAlaAT4) (Fig. 1). Also their flanking regions located up- and downstream of the two pairs of AlaAT sequences contained highly similar gene sequences. This indicates that the members within the AlaAT pairs originate from recent duplication events.
Sequence comparison of the four AlaATs from soybean. a Comparison of the intron and exon structure of the four AlaAT genomic sequences identified in soybean. The striking similarity between Glyma07g05130 and Glyma16g01630 or Glyma02g04320 and Glyma01g03260 from both coding and non-coding sequences is explained from genomic duplication events. Data are extracted from the Phytozome database at http://www.phytozome.net. b Alignment of the translated amino acid sequences from GmAlaAT1 (Glyma07g05130), GmAlaAT2 (Glyma02g04320), GmAlaAT3 (Glyma01g03260) and GmAlaAT4 (Glyma16g01630)
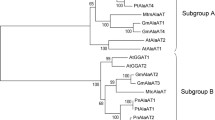
Classification of the AlaAT homologues was done by comparing full length protein sequences currently present in the public databases for AlaAT. The neighbour-joining phylogenetic tree that resulted from this analysis produced a clear separation into two subfamilies (Fig. 2), each of them containing two of the four homologues soybean genes that were identified. Glyma07g05130 (GmAlaAT1) and Glyma16g0163 (GmAlaAT4) clustered together in subfamily A, whereas Glyma02g04320 (GmAlaAT2) and Glyma01g03260 (GmAlaAT3) grouped together in subfamily B.
Phylogenetic tree of the AlaAT enzyme family. Unrooted neighbour-joining sequence comparison was performed using all fully sequenced coding sequences known to date for AlaAT genes in plants Chlamydomonas reinhardtii and Physcomitrella patens. Translated protein sequences were used for sequence alignment. A division into two subfamilies, A and B, can be observed. The full list with NCBI accession codes is given in “Materials and methods”. The four genes identified in soybean are marked in bold
Analysis of AlaAT in different organs of soybean
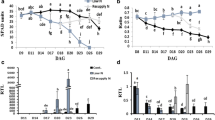
The distribution of AlaAT in adult soybean plants was analysed by measuring gene expression and by determining the activity of AlaAT (E.C.2.6.1.2.) in nodules, leaves and pods of both nodulated and non-nodulated plants. Furthermore, the amount of alanine was determined. Alanine levels varied strongly between the various organs, as well as between nodulated and non-nodulated plants (Fig. 3a). In non-nodulated plants, leaves contained the highest levels of alanine on a dry weight basis, whereas in nodulated plants the pods had the highest alanine content. Independent of the nodulation status, the level of alanine was lowest in roots.
Analysis of alanine content, AlaAT transcript levels and enzyme activity in various organs of soybean. a Alanine content in pods, leaves, roots and nodules (nod) of nodulated and non-nodulated soybean plants. b AlaAT enzyme activity measured in pods, leaves, roots and nodules (nod) of nodulated and non-nodulated soybean plants. c Semi-quantitive RT-PCR analysis of GmAlaAT transcript levels in pods, leaves, roots and nodules (nod) of nodulated and non-nodulated soybean plants. GmAlaAT-A represents transcript levels from the genes belonging to subfamily A (GmAlaAT1 and -4), whereas GmALaAT-B indicate transcripts from subfamily B (GmAlaAT2 and -3). Due to the very high sequence similarity, it was not possible to distinguish between individual genes within each subfamily. The constitutively expressed actin gene was used as loading control to normalise the samples. The bars indicate mean value ± SD (n = 4). Values that differ significantly according to a one-way ANOVA (P < 0.05) and Tukey posthoc test are marked with different letters. Small letters are used to indicate differences between tissues from either nodulated or non-nodulated plants. Capital letters indicate differences within the same kind of tissue from either nodulated or non-nodulated plants
The activity of AlaAT was tested by providing substrate that was specific to the AlaAT reaction (E.C.2.6.1.2) only. Activity appeared to be similar in all tissues except for the roots where AlaAT activity was about half that of the other organs (Fig. 3b). Nodulation had no influence on the activity of AlaAT in any of the organs tested.
Tissue-specific expression of the AlaAT homologues was determined by semi-quantitative reverse-transcription PCR (Fig. 3c). Due to the high sequence similarity between GmAlaAT1 and -4 and between GmAlaAT2 and -3, respectively, the obtained expression signals were likely to represent both members from either subfamily A (GmAlaAT1 (Glyma07g05130) and GmAlaAT4 (Glyma16g01630)) or B (GmAlaAT2 (Glyma02g04320) and GmAlaAT3 (Glyma01g03260)). Transcript levels of both AlaAT subfamilies were highest in nodules. Expression of subfamily B could also be detected in roots of both nodulated and non-nodulated plants, whereas transcript of subfamily A was well detected in roots of non-nodulated plants, and only very weak signals were obtained for these class A genes in roots from nodulated plants. In leaves and pods from nodulated and non-nodulated plants, the expression levels of all genes were only very low.
Effect of NO3 − or NH4 + fertilisation on the regulation of AlaAT in non-nodulated plants during waterlogging
To investigate the impact of nitrogen availability on the regulation AlaAT non-nodulated plants, transcript levels, enzyme activity and alanine content were investigated in the presence or absence of either nitrate or ammonium under both normoxia and waterlogging conditions. In roots of non-waterlogged plants without nodules, the content of alanine was relatively low and did not depend on the nitrogen availability of the plants (Fig. 4a). However, during waterlogging, the amount of alanine increased strongly from 0.74 before waterlogging to 23.97 nmol mg−1 DW during waterlogging in the absence of any N supply. In the presence of nitrate, the alanine content increased from 1.21 to 78.05 nmol mg−1 DW, whereas with ammonium the increase in alanine was even stronger, rising from 1.68 to 232.26 nmol mg−1 DW.
Hypoxic response of alanine accumulation and AlaAT enzyme activity and transcript level in relation to different nitrogen fertilisation conditions of non-nodulated and nodulated soybean plants. Alanine content (a, d), AlaAT activity (b, e) and AlaAT transcript levels as determined by semiquantitative RT-PCR (c, f) in roots or nodules of soybean plants without nodules (a–c) or with nodules (d–f). PCR conditions and sequence specificity are as described for Fig. 3. Material was analysed before the waterlogging treatment (C control) and after plants were waterlogged for 3 days (W). The bars indicate mean value ± SD as determined from four biological replicates. Values that differ significantly according to a one-way ANOVA (P < 0.05) and Tukey posthoc test are marked with different letters. Small letters are used to indicate differences induced by normoxia or hypoxia within the same nitrogen treatment. Capital letters indicate differences induced by the nitrogen treatment within either normoxic or hypoxic material
A similar tendency could be observed for the activity of AlaAT in roots of non-nodulated plants (Fig. 4b). No difference in AlaAT activity (E.C.2.6.1.2) was observed between control plants (before waterlogging) that were treated with various N supplies. During waterlogging, the activity of AlaAT increased by a factor of 2 during hypoxia to an activity of 0.9 μmol NADH min−1 mg protein in the absence of any extra N supply, whereas it increased to 1.20 μmol NADH min−1 mg protein in the presence of NO3 − and 1.64 μmol NADH min−1 mg protein in the presence of NH4 +. These data indicate that the external source of nitrogen affects alanine accumulation during waterlogging which correlates with the effect that the N supply has on the change in AlaAT activity (E.C.2.6.1.2) during waterlogging.
Semi-quantitative RT-PCR analysis revealed that the source of nitrogen that was supplied to non-nodulated plants strongly affected GmAlaAT expression levels (Fig. 4c). Transcript from genes belonging to subfamily A (GmAlaAT1 and -4) was lower in plants supplied with nitrate as compared to plants without special nitrogen fertilisation, whereas expression in ammonium-fed plants was much higher. In contrast, the transcript level from subfamily B genes (GmAlaAT2 and -3) was highest after feeding extra nitrate. Waterlogging strongly induced the expression of AlaAT subfamily A genes, independent of the nitrogen treatment the plant obtained. Different from that, the expression of genes from subfamily B decreased during waterlogging in both nitrate- and ammonium-treated plants, but increased in plants without nitrogen supplement.
Effect of NO3 − or NH4 + fertilisation on the regulation of AlaAT in nodulated plants during waterlogging
Similar to our analyses on non-nodulated plants, experiments were carried out on nodulated plants grown in the absence or presence of mineral N. Again, alanine accumulated in roots during waterlogging (Fig. 4d), but levels were 3–5 times less than in non-nodulated plants. In the absence of any external nitrogen supply, the amount of alanine increased during waterlogging by a factor of 11, rising from 0.60 to 6.85 nmol mg−1 DW. In the presence of NO3 − during waterlogging, alanine accumulation was approximately 24 times higher, rising from 0.74 to 17.57 nmol mg−1 DW, and when roots were supplemented with NH4 + alanine accumulated 22.3 times stronger (from 2,32 to 51.65 nmol mg−1 DW). In nodules, no significant change was observed in the amount of alanine except for nodules derived from plants fed with ammonium as these nodules showed a 17-time increase of the alanine concentration during waterlogging.
The change in activity of AlaAT in roots, as induced by waterlogging, was very similar between nodulated and non-nodulated plants. Upon waterlogging, AlaAT activity (E.C.2.6.1.2) in roots of both nodulated and non-nodulated plants increased two times in the absence of any external nitrogen source, whereas in the presence of NO3 − an increase of 2.7 times was observed, and after NH4 + was supplied AlaAT activity increased by 3.8 times (Fig. 4e).
The transcript level of subfamily A genes (AlaAT1 and -4) showed a strong increase during hypoxia in both roots and nodules independently of the nitrogen treatment given (Fig. 4f). In contrast, GmAlaAT subfamily B genes were expressed at a negligible level in all conditions tested.
Effect of hypoxia on AlaAT in soybean
It was suggested previously that AlaAT is likely involved in nitrogen mobilisation during the recovery period following waterlogging-induced hypoxic stress (de Sousa and Sodek 2003; Miyashita et al. 2007). Therefore, an experiment was performed to analyse changes of the alanine content, the activity of AlaAT and the changes in transcript abundance of the GmAlaAT homologues during and after a 3-day waterlogging treatment. Roots of both non-nodulated and nodulated plants were investigated before, during and after the 3-day period of waterlogging.
The content of alanine in root tissue increased during the 3-day period of hypoxia in both nodulated and non-nodulated plants. In the roots of non-nodulated plants, alanine increased 45 times and in root tissue of nodulated plants the increase was 4.6 times only (Fig. 5a). Independent of the nodulation status, the alanine values in roots recovered within the 3-day period of re-oxygenation to values close to those measured before the hypoxic treatment. The amount of alanine within nodules did not change during or after the waterlogging treatment.
Changes in alanine content, AlaAT activity and transcript levels in soybean tissues before and during a 3-day waterlogging treatment and subsequent recovery. a The amount of alanine as determined in roots and nodules of nodulated and non-nodulated soybean plants. b The activity of AlaAT as measured in extracts from nodulated and non-nodulated soybean plants. c Semiquantitive RT-PCR analysis of GmAlaAT transcript levels in roots and nodules of soybean. PCR conditions and sequence specificity are as described in Fig. 3. Samples were taken before waterlogging (C control), after 3 days of waterlogging (W), and 3 days after the waterlogging treatment was ceased (R recovery). The bars indicate mean value ± SD as determined from four biological replicates. Small letters indicate differences between the different treatments, C, W and R, within the same tissue. Capital letters indicate differences between nodulated and non-nodulated tissue within C, W or R treated samples
The changes in AlaAT activity (E.C.2.6.1.2) as induced by waterlogging and the subsequent recovery period (Fig. 5b) were similar to those changes observed for gene expression (Fig. 5c). In root tissue of non-nodulated plants, hypoxic conditions induced AlaAT activity by about two times, and a threefold increase was observed in roots from nodulated plants. During the recovery phase, the activity of AlaAT decreased slightly again, but even 3 days after the waterlogging treatment the activity of AlaAT remained about 1.5–1.9 times higher than the activity that was measured before the start of the experiment. Apparently, during re-oxygenation, the AlaAT protein stability is higher than the stability of the transcript. In nodules, the activity of AlaAT did change neither during nor after waterlogging.
Expression analysis revealed that in roots only the transcript from GmAlaAT subfamily A increased during waterlogging, and it declined to levels below the initial value during the recovery treatment. Moreover, in nodules, the expression of GmAlaAT subfamily A increased strongly during waterlogging, but in contrast to roots, the transcript did not decrease completely during the 3-day recovery period. Opposite to the expression changes observed in roots for GmAla1 and -4, the transcript level from GmAlaAT2 and -3 decreased during waterlogging.
Discussion
The regulation of AlaAT enzyme activity (E.C. 2.6.1.2) was investigated in soybean. First, the soybean genome sequencing data (Schmutz et al. 2010) were explored to identify AlaAT homologues encoded by the soybean genome. The analysis revealed the existence of four genes. Comparison between these sequences and homologues from other plant species revealed a subdivision into two subfamilies (Figs. 1, 2). The first subfamily A contained the soybean homologues GmAlaAT1 and -4, whereas both GmAlaAT2 and GmAlaAT3 were included in the subfamily B. The very high similarity in coding as well as non-coding sequences of the AlaAT genes indicated that the two members of each subfamily are derived from a recent genome duplication event.
In Arabidopsis, four AlaAT homologues were described also. Detailed functional analysis of these genes had revealed different substrate selectivity of the various enzymes that are encoded by these genes (Igarashi et al. 2003; Liepman and Olsen 2003). Two of them, AtAlaAT1 and AtAlaAT2, were annotated to encode true alanine aminotransferases (E.C. 2.6.1.2), whereas two others (AtGGAT1 and AtGGAT2) were shown to have glutamate:glyoxylate aminotransferase (GGAT) activity (EC. 2.6.1.4; Igarashi et al. 2003; Liepman and Olsen 2003). It is tempting to speculate that the two genes AlaAT2 and -3 from soybean that have highest sequence similarity to the Arabidopsis genes AtGGAT1 and -2 might share the same function in catalysing the glutamate:glyoxylate aminotransferase reaction (E.C.2.6.1.4) rather than being alanine aminotransferases (E.C.2.6.1.2). However, this prediction of enzyme function should be taken with care as it is based on functional analysis of very few members of the gene family only.
Using semi-quantitative RT-PCR, the changes in GmAlaAT gene expression were analysed. Due to the very high sequence similarity between the homologues from one subfamily, the expression signals were interpreted to represent the changes within the entire subfamily. Changes in the activity of AlaAT (E.C.2.6.1.2) in roots correlated well with the level of transcript of subfamily A (GmAlaAT1 and -4) (Fig. 4), indicating that these genes indeed encode for enzymes with true alanine aminotransferase activity (E.C.2.6.1.2) as was suggested earlier for Arabidopsis homologues from the same subfamily (Fig. 2; Miyashita et al. 2007). However, in other tissues than roots, the correlation between AlaAT activity and the expression of genes from subfamily A was not so obvious. For example, AlaAT activity was highest in pods and leaves, but transcript levels of each of the GmAlaAT homologues were very low. Apparently, under these steady state conditions, the activity of AlaAT enzyme was not determined by the transcript level which is easily explained when the AlaAT enzyme is stable under the given conditions.
A similar discrepancy between transcript levels and enzyme activity was observed when comparing samples taken during or after waterlogging of nodules (Figs. 4e, f, 5b, c). A comparable observation was described during the diurnal cycle for many more enzymes involved in primary carbohydrate metabolism (Blaesing et al. 2005). Probably, the stability of AlaAT protein can vary between tissues, or otherwise posttranscriptional regulation plays a role in the regulation of the activity of the enzyme. Henceforth, changes in gene expression are therefore always discussed in relation to the actual AlaAT enzyme activity.
It is well established that root AlaAT is highly activated upon hypoxia (de Sousa and Sodek 2003; Ricoult et al. 2006; Limami et al. 2008, Good et al. 2007; Miyashita et al. 2007; Beatty et al. 2009). Rocha et al. (2010) showed that the accumulation of alanine by AlaAT upon hypoxia occurred independent of the ability of the plant to fix nitrogen via symbiotic interaction with rhizobia. Even plants that were severely deprived of nitrogen were able to accumulate alanine during waterlogging. To investigate if an external nitrogen source has any effect on the activation of AlaAT during waterlogging, we investigated the expression and activity of AlaAT in plants that received various nitrogen supplements (Fig. 4). Indeed, in roots of both nodulated and non-nodulated plants, the activity of AlaAT as well as the accumulation of alanine was higher in plants that were supplied with NO3 − as compared the control plants. After NH4 + supply, AlaAT activity and alanine accumulation increased even further. Although the activity of AlaAT in both nodulated and non-nodulated plant was similar, non-nodulated plants had generally much higher amounts of alanine than nodulated plants. This is likely explained by the NO3 − fertilisation of the non-nodulated plants during growth resulting in the accumulation of NO3 − in root tissue followed by its mobilisation during hypoxia (Brandão and Sodek 2009). The high rate of alanine production during hypoxia in the presence of NH4 + was also observed by Vanlerberghe and Turpin (1990) for the green alga Selenastrum minutum. They suggested that the synthesis of alanine is part of an NH4 + detoxification mechanism without changing the energy or the redox potential of the cell. In either case, the high correlation between AlaAT activity and alanine accumulation strongly suggests that AlaAT is likely to be responsible for the accumulation of alanine during waterlogging.
It should be noted that the role of AlaAT is not limited to hypoxic conditions only. After the waterlogging treatment was ceased, the high amount of transcript from GmAlaAT subfamily A decreased rapidly, but the activity of the AlaAT enzyme remained at a level that was significantly above that found before waterlogging (Fig. 5). Probably, by catalysing the reversed reaction from alanine to pyruvate, the remaining activity of AlaAT can explain why the amount of alanine declined so rapidly during the recovery from waterlogging. de Sousa and Sodek (2003) came to a similar conclusion after measuring a detailed time-course of both AlaAT activity and the level of alanine in roots of soybean plants during waterlogging and on return to normoxia. They observed that much of the increase in AlaAT activity during waterlogging took place after most of the increase in alanine had occurred. On return to normoxia, the decrease in the level of alanine was very rapidly. It was concluded that the increase in AlaAT activity during waterlogging is particularly important to prepare the plant for the time that oxygen is available again. By catalysing the reaction from alanine to pyruvate, AlaAT would enable the tissue to re-utilise the alanine that accumulated during waterlogging. de Sousa and Sodek (2003) also observed high amounts of alanine being transported by the xylem from waterlogged roots to the normoxic shoot of the plant. This observation provides a further indication that alanine acts as an agent to recycle carbon and nitrogen efficiently within a waterlogged plant.
An alternative explanation for the role of AlaAT during hypoxia was raised by Drew (1997). He suggested that accumulation of alanine could improve tolerance to hypoxia via the activation of the glycolytic flux which increases the amount of ATP produced by the glycolytic pathway. However, it was argued that the involvement of AlaAT to drive glycolysis would be limited because NAD+ is not being regenerated from NADH, such as it is being done by the ethanol or lactate fermentation pathways. Alanine metabolism would therefore only be helpful for the production of ATP by glycolysis as long as NAD+ remains available. However, Rocha et al. (2010) described an extended reaction pathway which suggested that the production of alanine could be directly linked to the TCA cycle via ketoglutarate, which is produced concomitantly with alanine by AlaAT. This allows the plant to save NAD+ consumption in the TCA cycle by isocitrate dehydrogenase, but retains the production of ATP by oxoglutarate dehydrogenase.
The importance of AlaAT activity during hypoxia was further stressed by Zabalza et al. (2009). They described that the production of alanine from pyruvate is important for regulating the level of pyruvate as it was shown that increased concentrations of pyruvate lead to the activation of respiratory oxygen consumption, e.g. via the activation of the alternative oxidase in the mitochondria (Gupta et al. 2009). Especially during hypoxia, the consumption of oxygen should be reduced to a minimum. Therefore, mechanisms to prevent pyruvate accumulation are required. Temporal accumulation of alanine via AlaAT would serve that goal without the detrimental side effects that are induced by fermentation.
Summarising our data, evidence is provided here that the activity of AlaAT in soybean roots under hypoxia varies depending on the nitrogen source that is supplied to the plants with NH4 + inducing AlaAT activity more than NO3 −. The activation of AlaAT during waterlogging and the concomitant accumulation of alanine indicate a role of AlaAT in hypoxic metabolism. Nevertheless, it should be considered that the enzyme catalyses an equilibrium reaction and evidence is discussed that AlaAT also plays a role in alanine breakdown during the recovery from waterlogging stress.
References
Albrecht G, Kammerer S, Praznic W, Wiedenroth EM (1993) Fructan content of wheat seedlings (Triticum aestivum L.) under hypoxia and following re-aeration. New Phytol 123:471–476
Barret-Lennard EG, Leighton PD, Buwalda F et al (1988) Effects of growing wheat in hypoxic nutrient solutions and of subsequent transfer to aerated solutions. I. Growth and carbohydrate status of shoots and roots. Aust J Plant Physiol 15:585–598
Beatty PH, Shrawat AK, Carroll RT, Zhu T, Good AG (2009) Transcriptome analysis of nitrogen-efficient rice over-expressing alanine aminotransferase. Plant Biotechnol J 7:562–576
Bieleski RL, Turner NA (1966) Separation and estimation of amino acids in crude plant extracts by thin-layer electrophoresis and chromatography. Anal Biochem 17:278–293
Blaesing OE, Gibon Y, Guenther M, Hoehne M et al (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17:3257–3281
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brandão AD, Sodek L (2009) Nitrate uptake and metabolism by roots of soybean under oxygen deficiency. Braz J Plant Physiol 21:13–23
de Sousa CAF, Sodek L (2003) Alanine metabolism and alanine aminotransferase activity in soybean (Glycine max) during hypoxia of the root system and subsequent return to normoxia. Environ Exp Bot 50:1–8
Drew MC (1997) Oxygen deficiency and root metabolism: injury and acclimation under hypoxia and anoxia. Annu Rev Plant Physiol Plant Mol Biol 48:223–250
Fan TWM, Higashi RM, Lane AN (1988) An in vivo 1H and 31P NMR investigation of the effect of nitrate on hypoxic metabolism in maize roots. Arch Biochem Biophys 266:592–606
Good AG, Muench DG (1992) Purification and characterization of an aerobically induced alanine aminotransferase from barley roots. Plant Physiol 99:1520–1525
Good AG, Muench DG (1993) Long-term anaerobic metabolism in root-tissue (metabolic products of pyruvate metabolism). Plant Physiol 101:1163–1168
Good AG, Johnson SJ, De Pauw M, Carroll RT, Savidov N (2007) Engineering nitrogen use efficiency with alanine aminotransferase. Can J Bot 85:252–262
Gupta KJ, Zabalza A, Van Dongen JT (2009) Regulation of respiration when the oxygen availability changes. Physiol Plant 137:383–391
Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Hoagland DR, Arnon DI (1950) The water culture method of growing plants without soil. California Agricultural Experiment Station, Bulletin 347
Igarashi D, Miwa T, Seki M et al (2003) Identification of photorespiratory glutamate:glyoxylate aminotransferase (GGAT) gene in Arabidopsis. Plant J 33:975–987
Kikuchi H, Hirose S, Toki S, Akama K, Takaiwa F (1999) Molecular characterization of a gene for alanine aminotransferase from rice (Oryza sativa). Plant Mol Biol 39:149–159
Liepman AH, Olsen LJ (2003) Alanine aminotransferase homologs catalyze the glutamate:glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiol 131:215–227
Limami AM, Glévarec G, Ricoult C, Cliquet JB, Planchet E (2008) Concerted modulation of alanine and glutamate metabolism in young Medicago truncatula seedlings under hypoxic stress. J Exp Bot 59:2325–2335
Miyashita Y, Dolferus R, Ismond KP, Good AG (2007) Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana. Plant J 49:1108–1121
Muench DG, Good AG (1994) Hypoxically inducible barley alanine aminotransferase: cDNA cloning and expression analysis. Plant Mol Biol 24:417–427
Puiatti M, Sodek L (1999) Waterlogging affects nitrogen transport in the xylem of soybean. Plant Physiol Biochem 37:767–773
Ricoult C, Echeverria LO, Cliquet JB, Limami AM (2006) Characterization of alanine aminotransferase (AlaAT) multigene family and hypoxic response in young seedlings of the model legume Medicago truncatula. J Exp Bot 57:3079–3089
Rocha M, Licausi L, Araújo WL et al (2010) Glycolysis and the TCA-cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol 152:1501–1513
Schmutz J, Cannon SB, Schlueter J et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
van Dongen JT, Frohlich A, Ramirez-Aguilar SJ et al. (2009) Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of arabidopsis plants. Ann Bot 103:269–280
Vanlerberghe GC, Turpin DH (1990) Anaerobic metabolism in the N-limited green alga Selenastrum minutum II. Assimilation of ammonium by aerobic cells. Plant Physiol 94:1124–1130
Zabalza A, van Dongen JT, Froehlich A et al (2009) Regulation of respiration and fermentation to control the plant internal oxygen concentration. Plant Physiol 149:1087–1098
Acknowledgments
The Deutscher Akademischer Austausch Dienst (DAAD), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Deutsche Forschungs Gemeinschaft (SFB429) are kindly acknowledged for their financial support.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is published as part of the Special Issue on Plant Amino Acids.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Rocha, M., Sodek, L., Licausi, F. et al. Analysis of alanine aminotransferase in various organs of soybean (Glycine max) and in dependence of different nitrogen fertilisers during hypoxic stress. Amino Acids 39, 1043–1053 (2010). https://doi.org/10.1007/s00726-010-0596-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-010-0596-1