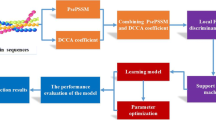
Summary.
Apoptosis proteins play an important role in the development and homeostasis of an organism. The accurate prediction of subcellular location for apoptosis proteins is very helpful for understanding the mechanism of apoptosis and their biological functions. However, most of the existing predictive methods are designed by utilizing a single classifier, which would limit the further improvement of their performances. In this paper, a novel predictive method, which is essentially a multi-classifier system, has been proposed by combing a dual-layer support vector machine (SVM) with multiple compositions including amino acid composition (AAC), dipeptide composition (DPC) and amphiphilic pseudo amino acid composition (Am-Pse-AAC). As a demonstration, the predictive performance of our method was evaluated on two datasets of apoptosis proteins, involving the standard dataset ZD98 generated by Zhou and Doctor, and a larger dataset ZW225 generated by Zhang et al. With the jackknife test, the overall accuracies of our method on the two datasets reach 94.90% and 88.44%, respectively. The promising results indicate that our method can be a complementary tool for the prediction of subcellular location.
Similar content being viewed by others
References
C Angulo X Parra A Català (2003) ArticleTitleK-SVCR. A support vector machine for multi-class classification Neurocomputing 55 57–77 Occurrence Handle10.1016/S0925-2312(03)00435-1
A Bairoch R Apweiler (2000) ArticleTitleThe SWISS-PROT protein sequence database and its supplement TrEMBL in 2000 Nucleic Acids Res 28 45–48 Occurrence Handle10592178 Occurrence Handle10.1093/nar/28.1.45 Occurrence Handle1:CAS:528:DC%2BD3cXhvVGqu7s%3D
M Bhasin GPS Raghava (2004) ArticleTitleESLpred: SVM-based method for subcellular localization of eukaryotic proteins using dipeptide composition and PSI-BLAST Nucleic Acids Res 32 W414–W419 Occurrence Handle15215421 Occurrence Handle10.1093/nar/gkh350 Occurrence Handle1:CAS:528:DC%2BD2cXlvFKnurk%3D
MPS Brown WN Grundy D Lin N Cristianini CW Sugnet TS Furey M Ares D Haussler (2000) ArticleTitleKnowledge-based analysis of microarray gene expression data by using support vector machines Proc Natl Acad Sci USA 97 262–267 Occurrence Handle10618406 Occurrence Handle10.1073/pnas.97.1.262 Occurrence Handle1:CAS:528:DC%2BD3cXjvVGjtw%3D%3D
A Bulashevska R Eils (2006) ArticleTitlePredicting protein subcellular locations using hierarchical ensemble of Bayesian classifiers based on Markov chains BMC Bioinformatics 7 298 Occurrence Handle16774677 Occurrence Handle10.1186/1471-2105-7-298 Occurrence Handle1:CAS:528:DC%2BD28XmtlWgtr4%3D
C Chen YX Tian XY Zou PX Cai JY Mo (2006a) ArticleTitleUsing pseudo-amino acid composition and support vector machine to predict protein structural class J Theor Biol 243 444–448 Occurrence Handle10.1016/j.jtbi.2006.06.025 Occurrence Handle1:CAS:528:DC%2BD28XhtFKlsL3N
C Chen XB Zhou YX Tian XY Zhou PX Cai (2006b) ArticleTitlePredicting protein structural class with pseudo-amino acid composition and support vector machine fusion network Anal Biochem 357 116–121 Occurrence Handle10.1016/j.ab.2006.07.022 Occurrence Handle1:CAS:528:DC%2BD28XpsVOgs78%3D
J Chen H Liu J Yang KC Chou (2007) ArticleTitlePrediction of linear B-cell epitopes using amino acid pair antigenicity scale Amino Acids 33 423–428 Occurrence Handle17252308 Occurrence Handle10.1007/s00726-006-0485-9 Occurrence Handle1:CAS:528:DC%2BD2sXpvVagsrc%3D
KC Chou (2001) ArticleTitlePrediction of protein cellular attributes using pseudo-amino acid composition Proteins Struct Funct Genet 43 246–255 Occurrence Handle11288174 Occurrence Handle10.1002/prot.1035 Occurrence Handle1:CAS:528:DC%2BD3MXjtFOls74%3D
KC Chou (2004) ArticleTitleReview: structural bioinformatics and its impact to biomedical science Curr Med Chem 11 2105–2134 Occurrence Handle15279552 Occurrence Handle1:CAS:528:DC%2BD2cXlslWltbw%3D
KC Chou (2005) ArticleTitleUsing amphiphilic pseudo amino acid composition to predict enzyme subfamily classes Bioinformatics 21 10–19 Occurrence Handle15308540 Occurrence Handle10.1093/bioinformatics/bth466 Occurrence Handle1:CAS:528:DC%2BD2MXisVWitw%3D%3D
Chou KC (2006) Frontiers in medicinal chemistry. In: Atta-ur-Rahman, Reitz AB (eds) Bentham Science Publishers, The Netherlands, pp 455–502
KC Chou YD Cai (2002) ArticleTitleUsing functional domain composition and support vector machines for prediction of protein subcellular location J Biol Chem 227 45765–45769 Occurrence Handle10.1074/jbc.M204161200 Occurrence Handle1:CAS:528:DC%2BD38XovFKjurg%3D
KC Chou YD Cai (2003) ArticleTitlePrediction and classification of protein subcellular location – sequence-order effect and pseudo amino acid composition J Cell Biochem 90 1250–1260 Occurrence Handle14635197 Occurrence Handle10.1002/jcb.10719 Occurrence Handle1:CAS:528:DC%2BD3sXpslSgsb4%3D
KC Chou YD Cai (2004a) ArticleTitlePredicting subcellular localization of proteins by hybridizing functional domain composition and pseudo-amino acid composition J Cell Biochem 91 1197–1203 Occurrence Handle10.1002/jcb.10790 Occurrence Handle1:CAS:528:DC%2BD2cXjt1yntrY%3D
KC Chou YD Cai (2004b) ArticleTitlePrediction of protein subcellular locations by GO-FunD-PseAA predictor Biochem Biophys Res Commun 320 1236–1239 Occurrence Handle10.1016/j.bbrc.2004.06.073 Occurrence Handle1:CAS:528:DC%2BD2cXls1eisL0%3D
KC Chou D Jones RL Heinrikson (1997) ArticleTitlePrediction of the tertiary structure and substrate binding site of caspase-8 FEBS Lett 419 49–54 Occurrence Handle9426218 Occurrence Handle10.1016/S0014-5793(97)01246-5 Occurrence Handle1:CAS:528:DyaK2sXnvFGlu74%3D
KC Chou HB Shen (2006a) ArticleTitlePredicting eukaryotic protein subcellular location by fusing optimized evidence-theoretic k-nearest neighbor classifiers J Proteome Res 5 1888–1897 Occurrence Handle10.1021/pr060167c Occurrence Handle1:CAS:528:DC%2BD28XmvVeitr0%3D
KC Chou HB Shen (2006b) ArticleTitleHum-PLoc: a novel ensemble classifier for predicting human protein subcellular localization Biochem Biophys Res Commun 347 150–157 Occurrence Handle10.1016/j.bbrc.2006.06.059 Occurrence Handle1:CAS:528:DC%2BD28Xmslyrsbc%3D
KC Chou HB Shen (2006c) ArticleTitlePredicting protein subcellular location by fusing multiple classifiers J Cell Biochem 99 517–527 Occurrence Handle10.1002/jcb.20879 Occurrence Handle1:CAS:528:DC%2BD28XhtVSktL3J
KC Chou HB Shen (2007a) ArticleTitleLarge-scale plant protein subcellular location prediction J Cell Biochem 100 665–678 Occurrence Handle10.1002/jcb.21096 Occurrence Handle1:CAS:528:DC%2BD2sXht1Slu7c%3D
KC Chou HB Shen (2007b) ArticleTitleEuk-mPLoc: a fusion classifier for large-scale eukaryotic protein subcellular location prediction by incorporating multiple sites J Proteome Res 6 1728–1734 Occurrence Handle1:CAS:528:DC%2BD2sXjs1SrsbY%3D Occurrence Handle10.1021/pr060635i
Chou KC, Shen HB (2007c) Review: recent progresses in protein subcellular location prediction. Anal Biochem doi: 10.1016/j.ab.2007.07.006
KC Chou HB Shen (2007d) ArticleTitleSignal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides Biochem Biophys Res Commun 357 633–640 Occurrence Handle10.1016/j.bbrc.2007.03.162 Occurrence Handle1:CAS:528:DC%2BD2sXkslCju78%3D
KC Chou HB Shen (2007e) ArticleTitleMemType-2L: a Web server for predicting membrane proteins and their types by incorporating evolution information through Pse-PSSM Biochem Biophys Res Commun 360 339–345 Occurrence Handle10.1016/j.bbrc.2007.06.027 Occurrence Handle1:CAS:528:DC%2BD2sXnslSqtLw%3D
KC Chou AG Tomasselli RL Heinrikson (2000) ArticleTitlePrediction of the tertiary structure of a caspase-9/inhibitor complex FEBS Lett 470 249–256 Occurrence Handle10745077 Occurrence Handle10.1016/S0014-5793(00)01333-8 Occurrence Handle1:CAS:528:DC%2BD3cXitVSrurw%3D
KC Chou CT Zhang (1995) ArticleTitleReview: prediction of protein structural classes Crit Rev Biochem Mol 30 275–349 Occurrence Handle10.3109/10409239509083488 Occurrence Handle1:CAS:528:DyaK2MXosFentb8%3D
C Cortes V Vapnik (1995) ArticleTitleSupport-vector networks Mach Learn 20 273–297
CHQ Ding I Dubchak (2001) ArticleTitleMulti-class protein fold recognition using support vector machines and neural networks Bioinformatics 17 349–358 Occurrence Handle11301304 Occurrence Handle10.1093/bioinformatics/17.4.349 Occurrence Handle1:CAS:528:DC%2BD3MXjvVegurg%3D
YS Ding TL Zhang KC Chou (2007) ArticleTitlePrediction of protein structure classes with pseudo amino acid composition and fuzzy support vector machine network Protein Peptide Lett 14 811–815 Occurrence Handle10.2174/092986607781483778 Occurrence Handle1:CAS:528:DC%2BD2sXhtlWiur7J
P Du Y Li (2006) ArticleTitlePrediction of protein submitochondria locations by hybridizing pseudo-amino acid composition with various physicochemical features of segmented sequence BMC Bioinformatics 7 518 Occurrence Handle17134515 Occurrence Handle10.1186/1471-2105-7-518 Occurrence Handle1:CAS:528:DC%2BD28XhtlWlsb7E
O Emanuelsson H Nielsen S Brunak G Heijne (2000) ArticleTitlePredicting subcellular localization of proteins based on their N-terminal amino acid sequence J Mol Biol 300 1005–1016 Occurrence Handle10891285 Occurrence Handle10.1006/jmbi.2000.3903 Occurrence Handle1:CAS:528:DC%2BD3cXks1OntrY%3D
ZP Feng (2002) ArticleTitleAn overview on predicting the subcellular location of a protein In Silico Biol 2 291–303 Occurrence Handle12542414 Occurrence Handle1:CAS:528:DC%2BD38Xpsl2lu7k%3D
A Garg M Bhasin GPS Raghava (2005) ArticleTitleSupport vector machine-based method for subcellular location of human proteins using amino acid compositions, their order and similarity search J Biol Chem 280 14427–14432 Occurrence Handle15647269 Occurrence Handle10.1074/jbc.M411789200 Occurrence Handle1:CAS:528:DC%2BD2MXjtFSmt7g%3D
Y Gao SH Shao X Xiao YS Ding YS Huang ZD Huang KC Chou (2005) ArticleTitleUsing pseudo amino acid composition to predict protein subcellular location: approached with Lyapunov index, bessel function, and Chebyshev filter Amino Acids 28 373–376 Occurrence Handle15889221 Occurrence Handle10.1007/s00726-005-0206-9 Occurrence Handle1:CAS:528:DC%2BD2MXlt1Kmurw%3D
YZ Guo M Li M Lu Z Wen K Wang G Li J Wu (2006) ArticleTitleClassifying G protein-coupled receptors and nuclear receptors based on protein power spectrum from fast Fourier transform Amino Acids 30 397–402 Occurrence Handle16773242 Occurrence Handle10.1007/s00726-006-0332-z Occurrence Handle1:CAS:528:DC%2BD28Xls1egs7o%3D
TP Hopp KR Woods (1981) ArticleTitlePrediction of protein antigenic determinants from amino acid sequences Proc Natl Acad Sci USA 78 3824–3828 Occurrence Handle6167991 Occurrence Handle10.1073/pnas.78.6.3824 Occurrence Handle1:CAS:528:DyaL3MXksF2lu7s%3D
SJ Hua ZR Sun (2001) ArticleTitleSupport vector machine approach for protein subcellular location prediction Bioinformatics 17 721–728 Occurrence Handle11524373 Occurrence Handle10.1093/bioinformatics/17.8.721 Occurrence Handle1:CAS:528:DC%2BD3MXntFKjsb0%3D
Y Huang Y Li (2004) ArticleTitlePrediction of protein subcellular locations using fuzzy k-NN method Bioinformatics 20 21–28 Occurrence Handle14693804 Occurrence Handle10.1093/bioinformatics/btg366 Occurrence Handle1:CAS:528:DC%2BD2cXns1Ol
H Lin QZ Li (2007a) ArticleTitlePredicting conotoxin superfamily and family by using pseudo amino acid composition and modified Mahalanobis discriminant Biochem Biophys Res Commun 354 548–551 Occurrence Handle10.1016/j.bbrc.2007.01.011 Occurrence Handle1:CAS:528:DC%2BD2sXhtlOgtLo%3D
H Lin QZ Li (2007b) ArticleTitleUsing pseudo amino acid composition to predict protein structural class: approached by incorporating 400 dipeptide components J Comput Chem 28 1463–1466 Occurrence Handle10.1002/jcc.20554 Occurrence Handle1:CAS:528:DC%2BD2sXlslSgs7w%3D
DQ Liu H Liu HB Shen J Yang KC Chou (2007) ArticleTitlePredicting secretory protein signal sequence cleavage sites by fusing the marks of global alignments Amino Acids 32 493–496 Occurrence Handle17103116 Occurrence Handle10.1007/s00726-006-0466-z Occurrence Handle1:CAS:528:DC%2BD2sXlsVGnsL8%3D
BW Matthews (1975) ArticleTitleComparison of predicted and observed secondary structure of T4 phage lysozyme Biochim Biophys Acta 405 442–451 Occurrence Handle1180967 Occurrence Handle1:CAS:528:DyaE2MXlslCksbk%3D
S Mondal R Bhavna R Mohan Babu S Ramakumar (2006) ArticleTitlePseudo amino acid composition and multi-class support vector machines approach for conotoxin superfamily classification J Theor Biol 243 252–260 Occurrence Handle16890961 Occurrence Handle10.1016/j.jtbi.2006.06.014 Occurrence Handle1:CAS:528:DC%2BD28XhtVygtbzM
K Nakai M Kanehisa (1992) ArticleTitleA knowledge base for predicting protein localization sites in eukaryotic cells Genomics 14 897–911 Occurrence Handle1478671 Occurrence Handle10.1016/S0888-7543(05)80111-9 Occurrence Handle1:CAS:528:DyaK3sXhs1Clsbw%3D
B Niu YD Cai WC Lu GY Zheng KC Chou (2006) ArticleTitlePredicting protein structural class with AdaBoost learner Protein Peptide Lett 13 489–492 Occurrence Handle10.2174/092986606776819619 Occurrence Handle1:CAS:528:DC%2BD28XlsVGqs7o%3D
KJ Park M Kanehisa (2003) ArticleTitlePrediction of protein subcellular locations by support vector machines using compositions of amino acids and amino acid pairs Bioinformatics 19 1656–1663 Occurrence Handle12967962 Occurrence Handle10.1093/bioinformatics/btg222 Occurrence Handle1:CAS:528:DC%2BD3sXnt1Gqu78%3D
ME Peter AE Heufelder MO Hengartner (1997) ArticleTitleAdvances in apoptosis research Proc Natl Acad Sci USA 94 12736–12737 Occurrence Handle9398063 Occurrence Handle10.1073/pnas.94.24.12736 Occurrence Handle1:CAS:528:DyaK2sXnvFantL0%3D
A Reinhardt T Hubbard (1998) ArticleTitleUsing neural networks for prediction of the subcellular location of proteins Nucleic Acids Res 26 2230–2236 Occurrence Handle9547285 Occurrence Handle10.1093/nar/26.9.2230 Occurrence Handle1:CAS:528:DyaK1cXjtFylsLw%3D
HB Shen KC Chou (2007a) ArticleTitleUsing ensemble classifier to identify membrane protein types Amino Acids 32 483–488 Occurrence Handle10.1007/s00726-006-0439-2 Occurrence Handle1:CAS:528:DC%2BD2sXlsVGnsLY%3D
HB Shen KC Chou (2007b) ArticleTitleHum-mPLoc: an ensemble classifier for large-scale human protein subcellular location prediction by incorporating samples with multiple sites Biochem Biophys Res Commun 355 1006–1011 Occurrence Handle10.1016/j.bbrc.2007.02.071 Occurrence Handle1:CAS:528:DC%2BD2sXivVahur0%3D
HB Shen KC Chou (2007c) ArticleTitleVirus-PLoc: a fusion classifier for predicting the subcellular localization of viral proteins within host and virus-infected cells Biopolymers 85 233–240 Occurrence Handle10.1002/bip.20640 Occurrence Handle1:CAS:528:DC%2BD2sXhvFWhs70%3D
HB Shen J Yang KC Chou (2007a) ArticleTitleEuk-PLoc: an ensemble classifier for large-scale eukaryotic protein subcellular location prediction Amino Acids 33 57–67 Occurrence Handle10.1007/s00726-006-0478-8 Occurrence Handle1:CAS:528:DC%2BD2sXotVWru7Y%3D
HB Shen J Yang KC Chou (2007b) ArticleTitleReview: methodology development for predicting subcellular localization and other attributes of proteins Expert Rev Proteomic 4 453–463 Occurrence Handle10.1586/14789450.4.4.453 Occurrence Handle1:CAS:528:DC%2BD2sXptFyrtLo%3D
JY Shi SW Zhang Q Pan YM Cheng J Xie (2007) ArticleTitlePrediction of protein subcellular localization by support vector machines using multi-scale energy and pseudo amino acid composition Amino Acids 33 69–74 Occurrence Handle17235454 Occurrence Handle10.1007/s00726-006-0475-y Occurrence Handle1:CAS:528:DC%2BD2sXotVWru7g%3D
XD Sun RB Huang (2006) ArticleTitlePrediction of protein structural classes using support vector machines Amino Acids 30 469–475 Occurrence Handle16622605 Occurrence Handle10.1007/s00726-005-0239-0 Occurrence Handle1:CAS:528:DC%2BD28Xls1ehu7c%3D
C Tanford (1962) ArticleTitleContribution of hydrophobic interactions to the stability of the globular conformation of proteins J Am Chem Soc 84 4240–4247 Occurrence Handle10.1021/ja00881a009 Occurrence Handle1:CAS:528:DyaF3sXkslOnsQ%3D%3D
Wang M, Yang J, Chou KC (2005) Using string kernel to predict signal peptide cleavage site based on subsite coupling model. Amino Acids (Erratum, ibid. 2005, 29: 301) 28, 395–402
Z Wen M Li Y Li Y Guo K Wang (2007) ArticleTitleDelaunay triangulation with partial least squares projection to latent structures: a model for G-protein coupled receptors classification and fast structure recognition Amino Acids 32 277–283 Occurrence Handle16729188 Occurrence Handle10.1007/s00726-006-0341-y Occurrence Handle1:CAS:528:DC%2BD2sXhtFyhtLY%3D
X Xiao S Shao Y Ding Z Huang Y Huang KC Chou (2005) ArticleTitleUsing complexity measure factor to predict protein subcellular location Amino Acids 28 57–61 Occurrence Handle15611847 Occurrence Handle10.1007/s00726-004-0148-7 Occurrence Handle1:CAS:528:DC%2BD2MXhsVKqsro%3D
X Xiao SH Shao YS Ding ZD Huang KC Chou (2006) ArticleTitleUsing cellular automata images and pseudo amino acid composition to predict protein subcellular location Amino Acids 30 49–54 Occurrence Handle16044193 Occurrence Handle10.1007/s00726-005-0225-6 Occurrence Handle1:CAS:528:DC%2BD28XhsFCksrk%3D
ZR Yang (2004) ArticleTitleBiological applications of support vector machines Brief Bioinform 5 328–338 Occurrence Handle15606969 Occurrence Handle10.1093/bib/5.4.328 Occurrence Handle1:CAS:528:DC%2BD2MXhsVejt7o%3D
CS Yu CJ Lin JK Huwang (2004) ArticleTitlePredicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions Protein Sci 13 1402–1406 Occurrence Handle15096640 Occurrence Handle10.1110/ps.03479604 Occurrence Handle1:CAS:528:DC%2BD2cXjsFKks74%3D
SW Zhang Q Pan HC Zhang ZC Shao JY Shi (2006) ArticleTitlePrediction protein homo-oligomer types by pseudo amino acid composition: approached with an improved feature extraction and naive Bayes feature fusion Amino Acids 30 461–468 Occurrence Handle16773245 Occurrence Handle10.1007/s00726-006-0263-8 Occurrence Handle1:CAS:528:DC%2BD28Xls1egsr0%3D
Zhang TL, Ding YS (2007) Using pseudo amino acid composition and binary-tree support vector machines to predict protein structural classes. Amino Acids, doi: 10.1007/s00726-007-0496-1
ZH Zhang ZH Wang YX Wang (2006a) ArticleTitlePrediction of the subcellular location of apoptosis-related proteins with encoding based on grouped weight for protein sequence Acta Biophys Sin 22 275–282 Occurrence Handle1:CAS:528:DC%2BD2sXhsVKjsLbF
ZH Zhang ZH Wang ZR Zhang YX Wang (2006b) ArticleTitleA novel method for apoptosis protein subcellular localization prediction combing encoding based on grouped weight and support vector machine FEBS Lett 580 6169–6174 Occurrence Handle10.1016/j.febslet.2006.10.017 Occurrence Handle1:CAS:528:DC%2BD28XhtFOhsrzN
GP Zhou (1998) ArticleTitleAn intriguing controversy over protein structural class prediction J Protein Chem 17 729–738 Occurrence Handle9988519 Occurrence Handle10.1023/A:1020713915365 Occurrence Handle1:CAS:528:DyaK1MXnslaltw%3D%3D
GP Zhou N Assa-Munt (2001) ArticleTitleSome insights into protein structural class prediction Proteins Struct Func Genet 44 57–59 Occurrence Handle10.1002/prot.1071 Occurrence Handle1:CAS:528:DC%2BD3MXktlSnsbk%3D
GP Zhou K Doctor (2003) ArticleTitleSubcellular location prediction of apoptosis proteins Proteins Struct Func Genet 50 44–48 Occurrence Handle10.1002/prot.10251 Occurrence Handle1:CAS:528:DC%2BD3sXlsVKmug%3D%3D
Author information
Authors and Affiliations
Additional information
Authors’ address: Xiao-Zou, School of Chemistry and Chemical Engineering, Sun Yat-Sen University, Guangzhou 510275, P. R. China
Rights and permissions
About this article
Cite this article
Zhou, XB., Chen, C., Li, ZC. et al. Improved prediction of subcellular location for apoptosis proteins by the dual-layer support vector machine. Amino Acids 35, 383–388 (2008). https://doi.org/10.1007/s00726-007-0608-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-007-0608-y