Summary.
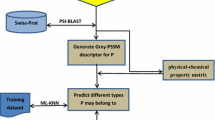
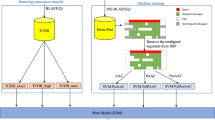
Predicting membrane protein type is both an important and challenging topic in current molecular and cellular biology. This is because knowledge of membrane protein type often provides useful clues for determining, or sheds light upon, the function of an uncharacterized membrane protein. With the explosion of newly-found protein sequences in the post-genomic era, it is in a great demand to develop a computational method for fast and reliably identifying the types of membrane proteins according to their primary sequences. In this paper, a novel classifier, the so-called “ensemble classifier”, was introduced. It is formed by fusing a set of nearest neighbor (NN) classifiers, each of which is defined in a different pseudo amino acid composition space. The type for a query protein is determined by the outcome of voting among these constituent individual classifiers. It was demonstrated through the self-consistency test, jackknife test, and independent dataset test that the ensemble classifier outperformed other existing classifiers widely used in biological literatures. It is anticipated that the idea of ensemble classifier can also be used to improve the prediction quality in classifying other attributes of proteins according to their sequences.
Similar content being viewed by others
References
B Alberts D Bray J Lewis M Raff K Roberts JD Watson (1994) Molecular biology of the cell EditionNumber3 Garland Publishing New York London
A Bairoch R Apweiler (2000) ArticleTitleThe SWISS-PROT protein sequence data bank and its supplement TrEMBL Nucleic Acids Res 25 31–36 Occurrence Handle10.1093/nar/25.1.31
YD Cai GP Zhou KC Chou (2003) ArticleTitleSupport vector machines for predicting membrane protein types by using functional domain composition Biophys J 84 3257–3263 Occurrence Handle12719255 Occurrence Handle1:CAS:528:DC%2BD3sXjvFGju7o%3D Occurrence Handle10.1016/S0006-3495(03)70050-2
J Cedano P Aloy JA P’erez-Pons E Querol (1997) ArticleTitleRelation between amino acid composition and cellular location of proteins J Mol Biol 266 594–600 Occurrence Handle9067612 Occurrence Handle10.1006/jmbi.1996.0804 Occurrence Handle1:CAS:528:DyaK2sXhslKksL4%3D
JJ Chou CT Zhang (1993) ArticleTitleA joint prediction of the folding types of 1490 human proteins from their genetic codons J Theor Biol 161 251–262 Occurrence Handle8331952 Occurrence Handle10.1006/jtbi.1993.1053 Occurrence Handle1:CAS:528:DyaK3sXkvV2ns7g%3D
KC Chou (1995) ArticleTitleA novel approach to predicting protein structural classes in a (20-1)-D amino acid composition space Proteins Struct Function Genet 21 319–344 Occurrence Handle10.1002/prot.340210406 Occurrence Handle1:CAS:528:DyaK2MXls12rsb0%3D
KC Chou (2001) ArticleTitlePrediction of protein cellular attributes using pseudo amino acid composition Proteins Struct Function Genet 43 246–255 Occurrence Handle10.1002/prot.1035 Occurrence Handle1:CAS:528:DC%2BD3MXjtFOls74%3D
KC Chou (2005) ArticleTitleReview: Progress in protein structural class prediction and its impact to bioinformatics and proteomics Curr Protein Pept Sci 6 423–436 Occurrence Handle16248794 Occurrence Handle10.2174/138920305774329368 Occurrence Handle1:CAS:528:DC%2BD2MXhtV2gt7zI
KC Chou DW Elrod (1999) ArticleTitlePrediction of membrane protein types and subcellular locations Proteins Struct Function Genet 34 137–153 Occurrence Handle10.1002/(SICI)1097-0134(19990101)34:1<137::AID-PROT11>3.0.CO;2-O Occurrence Handle1:CAS:528:DyaK1MXjtFGisg%3D%3D
KC Chou CT Zhang (1994) ArticleTitlePredicting protein folding types by distance functions that make allowances for amino acid interactions J Biol Chem 269 22014–22020 Occurrence Handle8071322 Occurrence Handle1:CAS:528:DyaK2cXlslCls7o%3D
KC Chou CT Zhang (1995) ArticleTitleReview: Prediction of protein structural classes Crit Rev Biochem Mol Biol 30 275–349 Occurrence Handle7587280 Occurrence Handle1:CAS:528:DyaK2MXosFentb8%3D
PY Chou (1989) Prediction of protein structural classes from amino acid composition GD Fasman (Eds) Prediction of protein structure and the principles of protein conformation Plenum Press New York 549–586
C Cortes V Vapnik (1995) ArticleTitleSupport vector networks Mach Learn 20 273–293
TM Cover PE Hart (1967) ArticleTitleNearest neighbour pattern classification IEEE Trans Inform Theory IT-13 21–27 Occurrence Handle10.1109/TIT.1967.1053964
ZP Feng (2001) ArticleTitlePrediction of the subcellular location of prokaryotic proteins based on a new representation of the amino acid composition Biopolymers 58 491–499 Occurrence Handle11241220 Occurrence Handle10.1002/1097-0282(20010415)58:5<491::AID-BIP1024>3.0.CO;2-I Occurrence Handle1:CAS:528:DC%2BD3MXisVSntb8%3D
ZP Feng (2002) ArticleTitleAn overview on predicting the subcellular location of a protein In Silico Biol 2 291–303 Occurrence Handle12542414 Occurrence Handle1:CAS:528:DC%2BD38Xpsl2lu7k%3D
Y Gao SH Shao X Xiao YS Ding YS Huang ZD Huang KC Chou (2005) ArticleTitleUsing pseudo amino acid composition to predict protein subcellular location: approached with Lyapunov index, Bessel function, and Chebyshev filter Amino Acids 28 373–376 Occurrence Handle15889221 Occurrence Handle10.1007/s00726-005-0206-9 Occurrence Handle1:CAS:528:DC%2BD2MXlt1Kmurw%3D
YZ Guo M Li M Lu Z Wen K Wang G Li J Wu (2006) ArticleTitleClassifying G protein-coupled receptors and nuclear receptors based on protein power spectrum from fast Fourier transform Amino Acids 30 397–402 Occurrence Handle16773242 Occurrence Handle10.1007/s00726-006-0332-z Occurrence Handle1:CAS:528:DC%2BD28Xls1egs7o%3D
Lodish H, Baltimore D, Berk A, Zipursky SL, Matsudaira P, Darnell J (1995) Mol Cell Biol, Chapter 3, 3rd ed. Scientific American Books, New York
RY Luo ZP Feng JK Liu (2002) ArticleTitlePrediction of protein strctural class by amino acid and polypeptide composition Eur J Biochem 269 4219–4225 Occurrence Handle12199700 Occurrence Handle10.1046/j.1432-1033.2002.03115.x Occurrence Handle1:CAS:528:DC%2BD38Xnt1eiur8%3D
PC Mahalanobis (1936) ArticleTitleOn the generalized distance in statistics Proc Natl Inst Sci India 2 49–55
KV Mardia JT Kent JM Bibby (1979) Multivariate analysis: Chapter 11: Discriminant analysis; Chapter 12: Multivariate analysis of variance; Chapter 13: cluster analysis Academic Press London 322–381
BW Matthews (1975) ArticleTitleComparison of the predicted and observed secondary structure of T4 phage lysozyme Biochim Biophys Acta 405 442–451 Occurrence Handle1180967 Occurrence Handle1:CAS:528:DyaE2MXlslCksbk%3D
H Nakashima K Nishikawa T Ooi (1986) ArticleTitleThe folding type of a protein is relevant to the amino acid composition J Biochem 99 152–162
YX Pan ZZ Zhang ZM Guo GY Feng ZD Huang L He (2003) ArticleTitleApplication of pseudo amino acid composition for predicting protein subcellular location: stochastic signal processing approach J Protein Chem 22 395–402 Occurrence Handle13678304 Occurrence Handle10.1023/A:1025350409648 Occurrence Handle1:CAS:528:DC%2BD3sXmsFejs7s%3D
KCS Pillai (1985) Mahalanobis D2 S Kotz NL Johnson (Eds) Encyclopedia of statistical sciences NumberInSeries5 Wiley New york 176–181
B Rost R Casadio P Fariselli C Sander (1995) ArticleTitleTransmembrane helices predicted at 95% accuracy Protein Sci 4 521–533 Occurrence Handle7795533 Occurrence Handle1:CAS:528:DyaK2MXksFyjsbY%3D Occurrence Handle10.1002/pro.5560040318
XD Sun RB Huang (2006) ArticleTitlePrediction of protein structural classes using support vector machines Amino Acids 30 469–475 Occurrence Handle16622605 Occurrence Handle10.1007/s00726-005-0239-0 Occurrence Handle1:CAS:528:DC%2BD28Xls1ehu7c%3D
M Wang J Yang GP Liu ZJ Xu KC Chou (2004) ArticleTitleWeighted-support vector machines for predicting membrane protein types based on pseudo amino acid composition Protein Eng Design Select 17 509–516 Occurrence Handle10.1093/protein/gzh061 Occurrence Handle1:CAS:528:DC%2BD2cXos1GisLY%3D
M Wang J Yang ZJ Xu KC Chou (2005) ArticleTitleSLLE for predicting membrane protein types J Theor Biol 232 7–15 Occurrence Handle15498588 Occurrence Handle10.1016/j.jtbi.2004.07.023 Occurrence Handle1:CAS:528:DC%2BD2cXovVKkur4%3D
X Xiao S Shao Y Ding Z Huang X Chen KC Chou (2005a) ArticleTitleAn application of gene comparative image for predicting the effect on replication ratio by HBV virus gene missense mutation J Theor Biol 235 555–565 Occurrence Handle10.1016/j.jtbi.2005.02.008 Occurrence Handle1:CAS:528:DC%2BD2MXltVelt7c%3D
X Xiao S Shao Y Ding Z Huang Y Huang KC Chou (2005b) ArticleTitleUsing complexity measure factor to predict protein subcellular location Amino Acids 28 57–61 Occurrence Handle10.1007/s00726-004-0148-7 Occurrence Handle1:CAS:528:DC%2BD2MXhsVKqsro%3D
X Xiao SH Shao YS Ding ZD Huang KC Chou (2006a) ArticleTitleUsing cellular automata images and pseudo amino acid composition to predict protein sub-cellular location Amino Acids 30 49–54 Occurrence Handle10.1007/s00726-005-0225-6 Occurrence Handle1:CAS:528:DC%2BD28XhsFCksrk%3D
X Xiao SH Shao ZD Huang KC Chou (2006b) ArticleTitleUsing pseudo amino acid composition to predict protein structural classes: approached with complexity measure factor J Comput Chem 27 478–482 Occurrence Handle10.1002/jcc.20354 Occurrence Handle1:CAS:528:DC%2BD28XitFyqsr4%3D
SW Zhang Q Pan HC Zhang ZC Shao JY Shi (2006) ArticleTitlePrediction protein homo-oligomer types by pseudo amino acid composition: approached with an improved feature extraction and naive Bayes feature fusion Amino Acids 30 461–468 Occurrence Handle16773245 Occurrence Handle10.1007/s00726-006-0263-8 Occurrence Handle1:CAS:528:DC%2BD28Xls1egsr0%3D
GP Zhou N Assa-Munt (2001) ArticleTitleSome insights into protein structural class prediction Proteins Struct Function Genet 44 57–59 Occurrence Handle10.1002/prot.1071 Occurrence Handle1:CAS:528:DC%2BD3MXktlSnsbk%3D
GP Zhou YD Cai (2006) ArticleTitlePredicting protease types by hybridizing gene ontology and pseudo amino acid composition Proteins Struct Function Bioinform 63 681–684 Occurrence Handle10.1002/prot.20898 Occurrence Handle1:CAS:528:DC%2BD28XksFaitb8%3D
GP Zhou K Doctor (2003) ArticleTitleSubcellular location prediction of apoptosis proteins Proteins Struct Function Genet 50 44–48 Occurrence Handle10.1002/prot.10251 Occurrence Handle1:CAS:528:DC%2BD3sXlsVKmug%3D%3D
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Shen, HB., Chou, KC. Using ensemble classifier to identify membrane protein types. Amino Acids 32, 483–488 (2007). https://doi.org/10.1007/s00726-006-0439-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-006-0439-2