Abstract
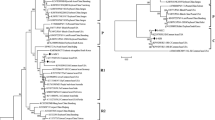
Tobacco vein banding mosaic virus (TVBMV) is of increasing importance in tobacco production. Knowledge of the genetic structure and variability of the virus population is vital for developing sustainable management. In this study, 24 new TVBMV isolates from Sichuan Province together with 46 previous isolates were studied based on their coat protein sequences. Two distinguishable clades were supported by phylogenetic analysis. The summary statistics PS, AI and MC showed a strong TVBMV-geography association between the isolates from Southwest China (SW) and Mainland China (MC). Further analysis indicated that the spatial genetic structure of TVBMV populations is likely to have been caused by natural selection. Phylogeographic analysis provided strong support for spatial diffusion pathways between the Southwest and Northwest tobacco-producing regions. The TVBMV CP gene was found to be under negative selection, and no significant positive selection of amino acids was detected in the SW group; however, the isolates of the MC group experienced significant positive selection pressure at the first and third amino acid sites of CP. This study suggests that natural selection and habitat heterogeneity are important evolutionary mechanisms affecting the genetic structure of the TVBMV population.


Similar content being viewed by others
References
Gibbs A, Ohshima K (2010) Potyviruses and the digital revolution. Annu Rev Phytopathol 48:205–223
Biebricher CK, Eigen M (2006) What is a Quasispecies? Curr Top Microbiol 299:1–31
Zhan J, McDonald BA (2004) The interaction among evolutionary forces in the pathogenic fungus Mycosphaerella graminicola. Fungal Genet Biol 41:590–599
Gao FL, Zou W, Xie LX (2017) Zhan JS (2017) Adaptive evolution and demographic history contribute to the divergent population genetic structure of Potato virus Y between China and Japan. Evol Appl 10:379–390
Thrall PH, Oakeshott JG, Fitt G, Southerton S, Burdon JJ, Sheppard A, Denison RF (2011) Evolution in agriculture: the application of evolutionary approaches to the management of biotic interactions in agro-ecosystems. Evol Appl 4:200–215
Pérez-Losada M, Arenas M, Galán JG, Palero F, González-Candelas F (2015) Recombination in viruses: mechanisms, methods of study, and evolutionary consequences. Infect Genet Evol 30:296–300
Revers F, García JA (2015) Molecular biology of potyviruses. Adv Virus Res 92:101–199
Yu XQ, Lan Y, Wang H, Liu J, Zhu X, Valkonen J, Li XD (2007) The complete genomic sequence of tobacco vein banding mosaic virus and its similarities with other potyviruses. Virus Genes 35:801–806
Rodamilans B, Valli A, Mingot A, San León D, Baulcombe D, López-Moya JJ, García JA (2015) RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the Potyviridae family. J Virol 89:6965–6967
Zhou GC, Wu XY, Zhang YM, Wu P, Wu XZ, Liu LW, Chen JQ (2014) A genomic survey of thirty soybean-infecting bean common mosaic virus (BCMV) isolates from China pointed BCMV as a potential threat to soybean production. Virus Res 191:125–133
Zhou GC, Shao ZQ, Ma FF, Wu P, Wu XY, Xie ZY, Chen JQ (2015) The evolution of soybean mosaic virus: an updated analysis by obtaining 18 new genomic sequences of Chinese strains/isolates. Virus Res 208:189–198
Lecoq H, Wipf Scheibel C, Chandeysson C, LêVan A, Fabre F, Desbiez C (2009) Molecular epidemiology of Zucchini yellow mosaic virus in France: an historical overview. Virus Res 141:190–200
Ohshima K, Yamaguchi Y, Hirota R, Hamamoto T, Tomimura K, Tan Z, Sano T, Azuhata F, Walsh JA, Fletcher J, Chen J, Gera A, Gibbs A (2002) Molecular evolution of Turnip mosaic virus: evidence of host adaptation, genetic recombination and geographical spread. J Gen Virol 83:1511–1521
Nguyen HD, Tran HTN, Ohshima K (2013) Genetic variation of the Turnip mosaic virus population of Vietnam: a case study of founder, regional and local influences. Virus Res 171:138–149
Habera LF, Berger PH, Reddick BB (1994) Molecular evidence from 3´-terminus sequence-analysis that tobacco vein-banding mosaic virus is a distinct member of the Potyvirus group. Arch Virol 138:27–38
Chin WT (1966) A survey of tobacco mosaic viruses in central Taiwan. J Agric Ass Chin 55:85–88
Dean CE, Clark F (1968) Vein-banding on tobacco in North Florida. Plant Dis Rep 52:887–889
Reddick BB, Collins-Shepard MH, Christie RG, Gooding GV (1992) A new virus-disease in North-America caused by Tobacco vein-banding mosaic virus. Plant Dis 76:856–859
Zhang CL, Gao R, Wang J, Zhang GM, Li XD, Liu HT (2011) Molecular variability of Tobacco vein banding mosaic virus populations. Virus Res 15:188–198
Hu XJ, Alexander VK, Celeste JB, Jim HL (2009) Sequence characteristics of potato virus Y recombinants. J Gen Virol 90:3033–3041
Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463
Nylander JAA (2008) MrModeltest v2.3. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Parker J, Rambaut A, Pybus OG (2008) Correlating viral phenotypes with phylogeny: Accounting for phylogenetic uncertainty. Infect Genet Evol 8:239–246
Hudson RR (2000) A new statistic for detecting genetic differentiation. Genetics 155:2011–2014
Kosakovsky Pond SL, Frost SDW, Grossman Z, Gravenor MB, Richman DD, Leigh Brown AJ (2006) Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput Biol 2:e62
Kosakovsky Pond SL, Murrell B, Fourment M, Frost SDW, Delport W, Scheffler K (2011) A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol 28:3033–3043
Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL (2012) Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8:e1002764
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24(8):1586–1591
Yang Z, Wong WSW, Nielsen R (2005) Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22(4):1107–1118
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973
Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV (2012) Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 29(9):2157–2167
Vaughan TG, Kühnert D, Popinga A, Welch D, Drummond AJ (2014) Efficient Bayesian inference under the structured coalescent. Bioinformatics 30(16):2272–2279
Drummond AJ, Rambaut A, Shapiro B, Pybus OG (2005) Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22(5):1185–1192
Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4(5):e88
Lemey P, Rambaut A, Drummond AJ, Suchard MA (2009) Bayesian phylogeography finds its roots. PloS Comput Biol 5:e1000520
Minin VN, Suchard MA (2008) Counting labeled transitions in continuous-time Markov models of evolution. J Math Biol 56:391–412
Traore O, Sorho F, Pinel A, Abubakar Z, Banwo O, Maley J et al (2010) Processes of diversification and dispersion of rice yellow mottle virus inferred from large-scale and high-resolution phylogeographical studies. Mol Ecol 14(7):2097–2110
Revell LJ, Harmon LJ, Collar DC (2008) Phylogenetic signal, evolutionary process and rate. Syst Biol 57:591–601
Caruso T, Chan Y, Lacap DC, Lau MC, McKay CP, Pointing SB (2011) Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J 5:1406–1413
Banke S, Lillemark MR, Gerstoft J, Obel N, Jørgensen LB (2009) Positive selection pressure introduces secondary mutations at Gag cleavage sites in Human immunodeficiency virus Type 1 harboring major protease resistance mutations. J Virol 83:8916–8924
Yu SF, Benton P, Bovee M, Sessions J, Lloyd RE (1995) Defective RNA replication by poliovirus mutants deficient in 2A protease cleavage activity. J Virol 69:247–252
Petrik K, Sebestyen E, Gell G, Balazs E (2010) Natural insertions within the N-terminal region of the coat protein of Maize dwarf mosaic potyvirus (MDMV) have an effect on the RNA stability. Virus Genes 40:135–139
Ullah Z, Chai B, Hammar S, Raccah B, Galon A, Grumet R (2003) Effect of substitution of the amino termini of coat proteins of distinct potyvirus species on viral infectivity and host specificity. Physiol Mol Plant Pathol 63(3):129–139
Feki S, Bouslama L (2008) Molecular phylogeny and genetic variability of the Potato virus Y (PVY) strains on the CP-encoding region. Ann Microbiol 58(3):433–438
Wang J, Whitlock MC (2003) Estimating effective population size and migration rates from genetic samples over space and time. Genetics 163:429–446
Gao FL, Liu XW, Du ZG, Han Hou H, Wang XY, Wang FL, Yang JG (2019) Bayesian phylodynamic analysis reveals the dispersal patterns of tobacco mosaic virus in China. Virology 528:110–117
Acknowledgements
We sincerely thank Dr. Fangluan Gao, Fujian Agriculture and Forestry University, China, for guiding the use of software.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Stephen John Wylie.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wei, S., He, X., Wang, D. et al. Genetic structure and variability of tobacco vein banding mosaic virus populations. Arch Virol 164, 2459–2467 (2019). https://doi.org/10.1007/s00705-019-04342-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04342-6