Abstract
A sensitive ratiometric fluorescence probe based on hybridization chain reaction (HCR) was constructed for sensitive detection of miRNA-21 by using G-triplex and silver nanocluster pairs (AgNC pairs) as an enzyme-free and label-free signal output group. miRNA-21 was used as the primer for the hybridization chain reaction of molecular beacon 1 (MB1) containing the locked G-triplex sequence and molecular beacon 2 (MB2) with intact AgNC pairs at the 5′ and 3′ end activation. The double-stranded product was obtained along with the opening of the G-triplex and the separation of the AgNC pairs. A detection limit of 67 pM and a linear detection range of 0.1–300 nM were obtained for miRNA-21 determination. The proposed strategy enabled the monitoring of miRNA-21 levels in at least three cell lines, indicating that it provided new ideas for detecting miRNA in real samples.
Graphical abstract

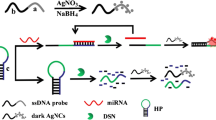
MB1 and MB2 contained the locked G-triplex sequence and silver nanocluster pairs (AgNC pairs), respectively. In the presence of target, the hybridization chain reaction (HCR) between MB1 and MB2 was initiated. At the same time, the locked G-triplex was released and combined to the dye thioflavin T (THT) to increase fluorescence, while the separation of the AgNC pairs caused the fluorescence to decrease. The double-stranded (ds) DNA product was generated to form a ratiometric signal to be detected.





Similar content being viewed by others
References
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18:997–1006. https://doi.org/10.1038/cr.2008.282
Cissell KA, Shrestha S, Deo SK (2007) MicroRNA detection: challenges for the analytical chemist. Anal Chem 79:4754–4761. https://doi.org/10.1021/ac0719305
Li R, Liu Q, Jin Y, Li B (2019) G-triplex/hemin DNAzyme: an ideal signal generator for isothermal exponential amplification reaction-based biosensing platform. Anal Chim Acta 1079:139–145. https://doi.org/10.1016/j.aca.2019.06.002
Wang S, Fu B, Wang J, Long Y, Zhang X, Peng S, Guo P, Tian T, Zhou X (2014) Novel amplex red oxidases based on noncanonical DNA structures: property studies and applications in microRNA detection. Anal Chem 86:2925–2930. https://doi.org/10.1021/ac402535a
Lu X, Li H, You J, Li W, Wang P, Li M, Dou S, Xi X (2018) Folding dynamics of parallel and anti-parallel G-triplex under the influence of proximal DNA. J Physl Chem B 122:9499–9506. https://doi.org/10.1021/acs.jpcb.8b08110
Li R, Liu Q, Jin Y, Li B (2020) Sensitive colorimetric determination of microRNA let-7a through rolling circle amplification and a peroxidase-mimicking system composed of trimeric G-triplex and hemin DNAzyme. Microchim Acta 187:139. https://doi.org/10.1007/s00604-019-4093-2
Linda C, Jussara A, Andrea G, Vittorio L, Ettore N, Michele P, Marco F, Antonio R, Claudio L (2014) G-triplex structure and formation propensity. Nucleic Acids Res 42:13393–13404. https://doi.org/10.1093/nar/gku1084
Frieg B, Gremer L, Heise H, Willbold D, Gohlke H (2020) Binding modes of thioflavin T and Congo red to the fibril structure of amyloid-β(1-42). Chem Commun 56:7589–7592. https://doi.org/10.1039/d0cc01161d
Zhou H, Wu ZF, Han QJ, Zhong HM, Peng JB, Li X, Fan XL (2018) Stable and label-free fluorescent probe based on G-triplex DNA and thioflavin T. Anal Chem 90:3220–3226. https://doi.org/10.1021/acs.analchem.7b04666
Guo R, Chen B, Li F, Weng S, Zheng Z, Chen M, Wu W, Lin X, Yang C (2018) Positive carbon dots with dual roles of nanoquencher and reference signal for the ratiometric fluorescence sensing of DNA. Sens Actuator B Chem 264:193–201. https://doi.org/10.1016/j.snb.2018.02.175
Guo Y, Chen Q, Qi Y, Xie Y, Qian H, Yao W, Pei R (2017) Label-free ratiometric DNA detection using two kinds of interaction responsive emission dyes. Biosens Bioelectron 87:320–324. https://doi.org/10.1016/j.bios.2016.08.041
Zhang Y, Guo S, Jiang Z, Mao G, Ji X, He Z (2018) Rox-DNA functionalized silicon nanodots for ratiometric detection of mercury ions in live cells. Anal Chem 90:9796–9804. https://doi.org/10.1002/adfm.201704092
Chen X, Xu K, Li J, Yang M, Li X, Chen Q, Lu C, Yang H (2020) Switch-conversional ratiometric fluorescence biosensor for miRNA detection. Biosens Bioelectron 155:112104–112109. https://doi.org/10.1016/j.bios.2020.112104
Ma L, Han X, Xia L, Kong R, Qu F (2018) G-triplex based molecular beacon for label-free fluorescence “turn-on” detection of bleomycin. Analyst 143:5474–5480. https://doi.org/10.1039/c8an01208c
Wu Z, Zhou H, He J, Li M, Ma X, Xue J, Li X, Fan X (2019) G-triplex based molecular beacon with duplex-specific nuclease amplification for specific detection of microRNA. Analyst 144:5201–5206. https://doi.org/10.1039/c9an01075k
Que H, Yan X, Guo B, Ma H, Wang T, Liu P, Gan X, Yan Y (2019) Terminal deoxynucleotidyl transferase and rolling circle amplification induced G-triplex formation: a label-free fluorescent strategy for DNA methyltransferase activity assay. Sens Actuators B chem 291:394–400. https://doi.org/10.1016/j.snb.2019.04.091
Wang K, He M, Zhai F, He R, Yu Y (2017) A label-free and enzyme-free ratiometric fluorescence biosensor for sensitive detection of carcinoembryonic antigen based on target-aptamer complex recycling amplification. Sens Actuators B Chem. 253:893–899. https://doi.org/10.1016/j.snb.2017.07.047
Ma J, Yin B, Ye B (2015) DNA template-regulated intergrowth of a fluorescent silver nanocluster emitter pair. RSC Adv 119:98467–98471. https://doi.org/10.1039/c5ra21159j
Yin B, Ma J, Le H, Wang S, Xu Z, Ye B (2019) A new mode to light up an adjacent DNA-scaffolded silver probe pair and its application for specific DNA detection. Chem Commun 100:15991–15994. https://doi.org/10.1039/c4cc07209j
Zhang M, Gao G, Ding Y, Deng C, Xiang J, Wu H (2019) A fluorescent aptasensor for the femtomolar detection of epidermal growth factor receptor-2 based on the proximity of G-rich sequences to Ag nanoclusters. Talanta 199:238–243. https://doi.org/10.1016/j.talanta.2019.02.014
Wang J, Zhang Z, Gao X, Lin X, Liu Y, Wang S (2019) A single fluorophore ratiometric nanosensor based on dual-emission DNA-templated silver nanoclusters for ultrasensitive and selective Pb2+ detection. Sens Actuators B Chem 282:712–718. https://doi.org/10.1016/j.snb.2018.11.121
Liu J, Lu Y, Feng L, Wang S, Zhang S, Zhu X, Sheng L, Zhang S, Zhang X (2017) Pinpoint the positions of single nucleotide polymorphisms by a nanocluster dimer. Anal Chem 89:2622–2627. https://doi.org/10.1021/acs.analchem.6b04981
Zhou W, Zhu J, Fan D, Teng Y, Zhu X, Dong S (2017) A multicolor chameleon DNA-templated silver nanocluster and its application for ratiometric fluorescence target detection with exponential signal response. Adv Funct Mater 27:1704092. https://doi.org/10.1002/adfm.201704092
Jiang H, Cui Y, Zhao T, Fu H, Koirala D, Punnoose JA, Kong D, Mao H (2015) Divalent cations and molecular crowding buffers stabilize G-triplex at physiologically relevant temperatures. Sci Rep 5:9255–9255. https://doi.org/10.1038/srep09255
Rajendran A, Endo M, Hidaka K, Teulade-Fichou MP, Mergny J, Sugiyama H (2015) Small molecule binding to a G-hairpin and a G-triplex: a new insight into anticancer drug design targeting G-rich regions. Chem Commun 44:9181–9184. https://doi.org/10.1039/c5cc01678a
Zhao L, Cao T, Zhou Q, Zhang X, Zhou Y, Yang L, Zhang X (2019) The exploration of a new stable G-triplex DNA and its novel function in electrochemical biosensing. Anal Chem 91:10731–10737. https://doi.org/10.1021/acs.analchem.9b02161
Liu Q, Sun X, Liu M, Jin Y, Li B (2020) G-triplex molecular beacon-based fluorescence biosensor for sensitive detection of small molecule-protein interaction via exonuclease III-assisted recycling amplification. Sens Actuators B Chem. 310:127804. https://doi.org/10.1016/j.snb.2020.127804
Wang Y, Wu Z, Liu Z (2013) Upconversion fluorescence resonance energy transfer biosensor with aromatic polymer nanospheres as the lable-free energy acceptor. Anal Chem 85:258–264. https://doi.org/10.1021/ac302659b
Xu S, Nie Y, Jing L, Wang J, Xu G, Wang W, Luo X (2018) Polydopamine nanosphere/gold nanocluster (Au NC)-based nanoplatform for dual color simultaneous detection of multiple tumor-related microRNAs with DNase-I-assisted target recycling amplification. Anal Chem 90:4039–4045. https://doi.org/10.1021/acs.analchem.7b05253
Zhang Y, Zhang C (2011) Sensitive detection of microRNA with isothermal amplification and a single-quantum-dot-based nanosensor. Anal Chem 84:224–231. https://doi.org/10.1021/ac202405q
Selcuklu SD, Donoghue MT, Spillane C (2009) MiR-21 as a key regulator of oncogenic processes. Biochem Soc Trans 37:918–925. https://doi.org/10.1042/bst0370918
Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY (2007) MiR-21-mediated tumor growth. Oncogene 26:2799–2803. https://doi.org/10.1038/sj.onc.1210083
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 21775132), the Natural Science Foundation of Hunan Province (No.2018JJ2388), Hunan 2011 Collaborative Innovation Center of Chemical Engineering & Technology with Environmental Benignity and Effective Resource Utilization, the project of innovation team of the Ministry of Education (IRT_17R90), and “1515” academic leader team program of Hunan Agricultural University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 191 kb)
Rights and permissions
About this article
Cite this article
Zhao, X., Wang, S., Zou, R. et al. An enzyme-free probe based on G-triplex assisted by silver nanocluster pairs for sensitive detection of microRNA-21. Microchim Acta 188, 55 (2021). https://doi.org/10.1007/s00604-020-04680-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-020-04680-2