Abstract
A method is described for the determination of microRNAs via two-stage signal enhancement. This is attained by combining hairpin (HP) assisted cascade isothermal amplification with light-up DNA-Ag nanoclusters. A rationally designed dual-functional HP is used, and microRNA-21 is chosen as a model analyte. At the first stage, upon the hybridization of the microRNA-21 with HP, microRNA recycling via polymerase-displacement reaction and a circulative nicking-replication process are achieved. This generates numerous G-abundant overhang DNA sequences. In the second stage, the above-released G-abundant overhang DNA sequences hybridize with the dark green Ag NCs, and this results in the appearance of bright red fluorescence. Thanks to the two signal enhancement processes, a linear dependence between the fluorescence intensity at 616 nm and the concentration of microRNA-21 is obtained in the range from 1 pM to 20 pM with a detection limit of 0.7 pM. The strategy clearly discriminates between perfectly-matched and mismatched targets. The method was applied to the determination of microRNA-21 in a spiked serum sample.

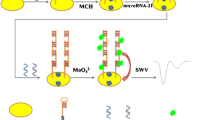
Schematic representation of microRNA detection by integrating hairpin assisted cascade isothermal amplification with light-up DNA Ag nanoclusters. With microRNA, G-abundant overhang DNA sequences from amplification reaction hybridize with dark green Ag nanoclusters to produce a concentration-dependent bright red fluorescence






Similar content being viewed by others
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. https://doi.org/10.1016/s0092-8674(04)00045-5
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531. https://doi.org/10.1038/nrg1379
Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM (2003) Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113(1):25–36. https://doi.org/10.1016/s0092-8674(03)00231-9
Wang Y, Keys DN, Au-Young JK, Chen C (2009) MicroRNAs in embryonic stem cells. J Cell Physiol 218(2):251–255. https://doi.org/10.1002/jcp.21607
Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G (2003) Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 9(2):180–186. https://doi.org/10.1261/rna.2141503
Xu P, Vernooy SY, Guo M, Hay BA (2003) The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol 13(9):790–795. https://doi.org/10.1016/s0960-9822(03)00250-1
Li C, Feng Y, Coukos G, Zhang L (2009) Therapeutic microRNA strategies in human cancer. AAPS J 11:747–757. https://doi.org/10.1208/s12248-009-9145-9
Dong H, Lei J, Ding L, Wen Y, Ju H, Zhang X (2013) MicroRNA: function, detection, and bioanalysis. Chem Rev 113(8):6207–6233. https://doi.org/10.1021/cr300362f
Yin JQ, Zhao RC, Morris KV (2008) Profiling microRNA expression with microarrays. Trends Biotechnol 26(2):70–76. https://doi.org/10.1016/j.tibtech.2007.11.007
Lee YS, Dutta A (2009) MicroRNAs in cancer. Annu Rev Pathol 4:199–227. https://doi.org/10.1146/annurev.pathol.4.110807.092222
Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T (2008) Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A 105(10):3903–3908. https://doi.org/10.1073/pnas.0712321105
Kosaka N, Iguchi H, Ochiya T (2010) Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 101(10):2087–2092. https://doi.org/10.1111/j.1349-7006.2010.01650.x
Grosshans H, Filipowicz W (2008) Molecular biology: the expanding world of small RNAs. Nature 451(7177):414–416. https://doi.org/10.1038/451414a
Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW (2006) Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature 442(7098):82–85. https://doi.org/10.1038/nature04836
Cissell KA, Shrestha S, Deo SK (2007) MicroRNA detection: challenges for the analytical chemist. Anal Chem 79(13):4754–4761. https://doi.org/10.1021/ac0719305
Válóczi A, Hornyik C, Varga N, Burgyán J, Kauppinen S, Havelda Z (2004) Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res 32(22):e175. https://doi.org/10.1093/nar/gnh171
Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B (2010) A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res 38(7):e98. https://doi.org/10.1093/nar/gkp1235
Jinek M, Doudna JA (2009) A three-dimensional view of the molecular machinery of RNA interference. Nature 457(7228):405–412. https://doi.org/10.1038/nature07755
Yun S, Kim WK, Kwon Y, Jang M, Bauer S, Kim H (2018) Survivin is a novel transcription regulator of KIT and is downregulated by miRNA-494 in gastrointestinal stromal tumors. Int J Cancer 142(10):2080–2093. https://doi.org/10.1002/ijc.31235
Shen Y, Zheng KX, Duan D, Jiang L, Li J (2012) Label-free microRNA profiling not biased by 3′ end 2'-O-methylation. Anal Chem 84(15):6361–6365. https://doi.org/10.1021/ac301360z
Ludlow AT, Robin JD, Sayed M, Litterst CM, Shelton DN, Shay JW, Wright WE (2014) Quantitative telomerase enzyme activity determination using droplet digital PCR with single cell resolution. Nucleic Acids Res 42(13):e104. https://doi.org/10.1093/nar/gku439
Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA (2007) Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res 67(6):2456–2468. https://doi.org/10.1158/0008-5472.CAN-06-2698
Miao X, Cheng Z, Ma H, Li Z, Xue N, Wang P (2018) Label-free platform for microRNA detection based on the fluorescence quenching of positively charged gold nanoparticles to silver nanoclusters. Anal Chem 90(2):1098–1103. https://doi.org/10.1021/acs.analchem.7b01991
Bi S, Ye J, Dong Y, Li H, Cao W (2016) Target-triggered cascade recycling amplification for label-free detection of microRNA and molecular logic operations. Chem Commun (Camb) 52(2):402–405. https://doi.org/10.1039/c5cc07046e
Jin J, Vaud S, Zhelkovsky AM, Posfai J, McReynolds LA (2016) Sensitive and specific miRNA detection method using SplintR ligase. Nucleic Acids Res 44(13):e116. https://doi.org/10.1093/nar/gkw399
Schneider N, Meier M (2017) Efficient in situ detection of mRNAs using the Chlorella virus DNA ligase for padlock probe ligation. RNA 23(2):250–256. https://doi.org/10.1261/rna.057836.116
He Y, Yang X, Yuan R, Chai Y (2017) “Off’ to “on” surface-enhanced raman spectroscopy platform with padlock probe-based exponential rolling circle amplification for ultrasensitive detection of microRNA 155. Anal Chem 89(5):2866–2872. https://doi.org/10.1021/acs.analchem.6b04082
Deng R, Zhang K, Sun Y, Ren X, Li J (2017) Highly specific imaging of mRNA in single cells by target RNA-initiated rolling circle amplification. Chem Sci 8(5):3668–3675. https://doi.org/10.1039/C7SC00292K
Wang M, Zhou Y, Yin H, Jiang W, Wang H, Ai S (2018) Signal-on electrochemiluminescence biosensor for microRNA-319a detection based on two-stage isothermal strand-displacement polymerase reaction. Biosens Bioelectron 107:34–39. https://doi.org/10.1016/j.bios.2018.02.015
Ou S, Xu T, Liu X, Yu XY, Li R, Deng J, Yuan JX, Chen YM (2018) Rapid and ultrasensitive detection of microRNA based on strand displacement amplification-mediated entropy-driven circuit reaction. Sensors Actuators B Chem 255(3):3057–3063. https://doi.org/10.1016/j.snb.2017.09.130
Zhang J, Li C, Zhi X, Ramón GA, Liu Y, Zhang C (2016) Hairpin DNA-templated silver nanoclusters as novel beacons in strand displacement amplification for microRNA detection. Anal Chem 88(2):1294–1302. https://doi.org/10.1021/acs.analchem.5b03729
Reid MS, Le XC, Zhang H (2018) Exponential isothermal amplification of nucleic acids and assays for proteins, cells, small molecules, and enzyme activities: an EXPAR example. Angew Chem Int Ed Engl 57(37):11856–11866. https://doi.org/10.1002/anie.201712217
Chen J, An T, Ma Y, Situ B, Chen D, Xu Y (2018) Isothermal amplification on a structure-switchable symmetric toehold dumbbell-template: a strategy enabling microRNA analysis at the single-cell level with ultrahigh specificity and accuracy. Anal Chem 90(1):859–865. https://doi.org/10.1021/acs.analchem.7b03713
Zhou X, Liang Y, Xu Y, Lin X, Chen J, Ma Y, Zhang L, Chen D, Song F, Dai Z, Zou X (2016) Triple cascade reactions: an ultrasensitive and specific single tube strategy enabling isothermal analysis of microRNA at sub-attomole level. Biosens Bioelectron 80:378–384. https://doi.org/10.1016/j.bios.2016.01.059
Duan R, Zuo X, Wang S, Quan X, Chen D, Chen Z, Jiang L, Fan C, Xia F (2014) Quadratic isothermal amplification for the detection of microRNA. Nat Protoc 9(3):597–607. https://doi.org/10.1038/nprot.2014.036
Duan R, Zuo X, Wang S, Quan X, Chen D, Chen Z, Jiang L, Fan C, Xia F (2013) Lab in a tube: ultrasensitive detection of microRNAs at the single-cell level and in breast cancer patients using quadratic isothermal amplification. J Am Chem Soc 135(12):4604–4607. https://doi.org/10.1021/ja311313b
Ma W, Fu P, Sun M, Xu L, Kuang H, Xu C (2017) Dual quantification of microRNAs and telomerase in living cells. J Am Chem Soc 139(34):11752–11759. https://doi.org/10.1021/jacs.7b03617
Hu J, Wang ZY, Li CC, Zhang CY (2017) Advances in single quantum dot-based nanosensors. Chem Commun (Camb) 53(100):13284–13295. https://doi.org/10.1039/C7CC07752A
Zhang Y, Zhang CY (2012) Sensitive detection of microRNA with isothermal amplification and a single-quantum-dot-based nanosensor. Anal Chem 84(1):224–231. https://doi.org/10.1021/ac202405q
Li J, Zhong X, Cheng F, Zhang JR, Jiang LP, Zhu JJ (2012) One-pot synthesis of aptamer-functionalized silver nanoclusters for cell-type-specific imaging. Anal Chem 84(9):4140–4146. https://doi.org/10.1021/ac3003402
Zhou WJ, Zhu JB, Fan DQ, Teng Y, Zhu XQ, Dong SJ (2017) A multicolor chameleon DNA-templated silver nanocluster and its application for ratiometric fluorescence target detection with exponential signal response. Adv Funct Mater 27(46):1704092. https://doi.org/10.1002/adfm.201704092
Guo W, Yuan J, Dong Q, Wang E (2010) Highly sequence-dependent formation of fluorescent silver nanoclusters in hybridized DNA duplexes for single nucleotide mutation identification. J Am Chem Soc 132(3):932–934. https://doi.org/10.1021/ja907075s
de Souza N (2007) All that glitters but does not blink. Nat Methods 4:540. https://doi.org/10.1038/nmeth0707-540
Li B, Xu L, Chen Y, Zhu W, Shen X, Zhu C, Luo J, Li X, Hong J, Zhou X (2017) Sensitive and label-free fluorescent detection of transcription factors based on DNA-Ag nanoclusters molecular beacons and exonuclease III-assisted signal amplification. Anal Chem 89(14):7316–7323. https://doi.org/10.1021/acs.analchem.7b00055
Liu G, Li J, Feng DQ, Zhu JJ, Wang W (2017) Silver nanoclusters beacon as stimuli-responsive versatile platform for multiplex DNAs detection and aptamer-substrate complexes sensing. Anal Chem 89(1):1002–1008. https://doi.org/10.1021/acs.analchem.6b04362
Chen A, Ma S, Zhuo Y, Chai Y, Yuan R (2016) In situ electrochemical generation of electrochemiluminescent silver naonoclusters on target-cycling synchronized rolling circle amplification platform for microRNA detection. Anal Chem 88(6):3203–3210. https://doi.org/10.1021/acs.analchem.5b04578
Ma JL, Yin BC, Wu X, Ye BC (2016) Copper-mediated DNA-scaffolded silver nanocluster on-off switch for detection of pyrophosphate and alkaline phosphatase. Anal Chem 88(18):9219–9225. https://doi.org/10.1021/acs.analchem.6b02465
Ma JL, Yin BC, Ye BC (2016) A versatile proximity-dependent probe based on light-up DNA-scaffolded silver nanoclusters. Analyst 141(4):1301–1306. https://doi.org/10.1039/C5AN02446C
Ma JL, Yin BC, Ye BC (2015) DNA template-regulated intergrowth of a fluorescent silver nanocluster emitter pair. RSC Adv 5:98467–98471. https://doi.org/10.1039/C5RA21159J
Petty JT, Giri B, Miller IC, Nicholson DA, Sergev OO, Banks TM, Story SP (2013) Silver clusters as both chromophoric reporters and DNA ligands. Anal Chem 85(4):2183–2190. https://doi.org/10.1021/ac303531y
Obliosca JM, Liu C, Yeh HC (2013) Fluorescent silver nanoclusters as DNA probes. Nanoscale 5(18):8443–8461. https://doi.org/10.1039/C3NR01601C
Yeh HC, Sharma J, Han JJ, Martinez JS, Werner JH (2010) A DNA--silver nanocluster probe that fluoresces upon hybridization. Nano Lett 10(8):3106–3110. https://doi.org/10.1021/nl101773c
Petty JT, Zheng J, Hud NV, Dickson RM (2004) DNA-templated Ag nanocluster formation. J Am Chem Soc 126(16):5207–5212. https://doi.org/10.1021/ja031931o
Richards CI, Choi S, Hsiang JC, Antoku Y, Vosch T, Bongiorno A, Tzeng YL, Dickson RM (2008) Oligonucleotide-stabilized Ag nanocluster fluorophores. J Am Chem Soc 130(15):5038–5039. https://doi.org/10.1021/ja8005644
Ying ZM, Wu Z, Tu B, Tan W, Jiang JH (2017) Genetically encoded fluorescent RNA sensor for ratiometric imaging of microRNA in living tumor cells. J Am Chem Soc 139(29):9779–9782. https://doi.org/10.1021/jacs.7b04527
Yi JT, Chen TT, Huo J, Chu X (2017) Nanoscale zeolitic imidazolate framework-8 for ratiometric fluorescence imaging of microRNA in living cells. Anal Chem 89(22):12351–12359. https://doi.org/10.1021/acs.analchem.7b03369
Acknowledgments
This work was supported by the Natural Science Foundation of China (21461002, 21241005 and 21562002), Key Laboratory of Jiangxi University for Functional Materials Chemistry (FMC15104), Jiangxi Provincial Department of Education (GJJ170826 and JXJG-14-6), and Gannan Normal University (16zb19).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 458 kb)
Rights and permissions
About this article
Cite this article
Li, M., Xu, X., Zhou, Z. et al. Label-free detection of microRNA: two-stage signal enhancement with hairpin assisted cascade isothermal amplification and light-up DNA-silver nanoclusters. Microchim Acta 187, 141 (2020). https://doi.org/10.1007/s00604-019-4094-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-4094-1