Abstract
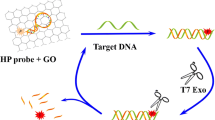
A fluorometric method is described for the determination of DNA. It involves the use of graphene oxide (GO), exonuclease III (Exo III), and two specially designed fluorophore-labeled hairpin probes (HP1 and HP2). Different from other GO-based DNA assays, the method takes advantage of the distinct binding abilities of GO with hairpin DNA probes and single nucleotides. GO serves as a strong quencher for fluorescent labels to ensure a very low background signal. Two reaction cycles mediated by Exo III are employed to enhance the signals. The combination of GO-induced quenching and Exo III-mediated dual regeneration of analytes leads to a detection limit as low as 1 pM for the model analyte human hemochromatosis protein (HFE) gene. The method is also applicable for the determination of HFE gene spiked into fetal bovine serum.

Schematic representation of a GO-based, Exo III-assisted method for dual-signal amplified detection of DNA, for which human haemochromatosis protein (HFE) gene is designed as the model target. The assay involves graphene oxide (GO), exonuclease (Exo III), and two specially designed, fluorophore-labelled hairpin probes (HP1 and HP2).




Similar content being viewed by others
References
Zhao X, Tapec-Dytioco R, Tan W (2013) Ultrasensitive DNA detection using highly fluorescent bioconjugated nanoparticles. J Am Chem Soc 125:11474–11475
Gerasimova YV, Kolpashchikov DM (2014) Enzyme-assisted target recycling (EATR) for nucleic acid detection. Chem Soc Rev 43:6405–6438
Huang J, Ye L, Gao X, Li H, Xu J, Li Z (2015) Molybdenum disulfide-based amplified fluorescence DNA detection using hybridization chain reactions. J Mater Chem B 3:2395–2401
Huang J, Su X, Li Z (2012) Enzyme-free and amplified fluorescence DNA detection using bimolecular beacons. Anal Chem 84:5939–5943
Huang J, Su X, Li Z (2014) Enzyme-and label-free amplified fluorescence DNA detection using hairpin probes and SYBR green I. Sensors Actuators B Chem 200:117–122
Huang J, Gao X, Jia J, Kim J, Li Z (2014) Graphene oxide-based amplified fluorescent biosensor for Hg2+ detection through hybridization chain reactions. Anal Chem 86:3209–3215
Zou L, Ling L (2018) Ultrasensitive detection of HIV DNA with polymerase chain reaction–dynamic light scattering. Anal Chem 90:13373–13377
Wee EJ, Shiddiky MJ, Brown MA, Trau M (2012) Electrochemical detection of single DNA base changes via ligase chain reaction. Chem Commun 48:12014–12016
Russell C, Welch K, Jarvius J, Cai Y, Brucas R, Nikolajeff F, Svedlindh P, Nilsson M (2014) Gold nanowire based electrical DNA detection using rolling circle amplification. ACS Nano 8:1147–1153
Park BH, Oh SJ, Jung JH, Choi G, Seo JH, Lee EY, Seo TS (2017) An integrated rotary microfluidic system with DNA extraction, loop-mediated isothermal amplification, and lateral flow strip based detection for point-of-care pathogen diagnostics. Biosens Bioelectron 91:334–340
Lei J, Ju H (2012) Signal amplification using functional nanomaterials for biosensing. Chem Soc Rev 41:2122–2134
Yang X, Tang Y, Mason SD, Chen J, Li F (2016) Enzyme-powered three-dimensional DNA nanomachine for DNA walking, payload release, and biosensing. ACS Nano 10:2324–2330
Deng X, Wang C, Wang S, Gao Y, Li J, Wen W, Zhang X (2018) Applying strand displacement amplification to quantum dots-based fluorescent lateral flow assay strips for HIV-DNA detection. Biosens Bioelectron 105:211–217
Fortunati S, Rozzi A, Curti F, Giannetto M, Corradini R, Careri M (2019) Novel amperometric genosensor based on peptide nucleic acid (PNA) probes immobilized on carbon nanotubes-screen printed electrodes for the determination of trace levels of non-amplified DNA in genetically modified (GM) soy. Biosens Bioelectron 129:7–14
Huang J, Zheng Q, Kim J, Li Z (2013) A molecular beacon and graphene oxide-based fluorescent biosensor for Cu2+ detection. Biosens Bioelectron 43:379–383
Huang J, Wang Z, Kim J, Su X, Li Z (2015) Detecting arbitrary DNA mutations using graphene oxide and ethidium bromide. Anal Chem 87:12254–12261
Gupta A, Sakthivel T, Seal S (2015) Recent development in 2D materials beyond graphene. Prog. Mater Sci 73:44–126
Lu C, Yang H, Zhu C, Chen X, Chen G (2009) A graphene platform for sensing biomolecules. Angew Chem 121:4879–4881
Kim J, Park S, Min D (2017) Emerging approaches for graphene oxide biosensor. Anal Chem 89:232–248
Iwe IA, Li Z, Huang J (2019) A dual-cycling fluorescent scheme for ultrasensitive DNA detection through signal amplification and target regeneration. Analyst 144:2649–2655
Cui L, Ke G, Zhang WY, Yang CJ (2011) A universal platform for sensitive and selective colorimetric DNA detection based on Exo III assisted signal amplification. Biosens Bioelectron 26:2796–2800
He S, Song B, Li D, Zhu C, Qi W, Wen Y, Wang L, Song S, Fang H, Fan C (2010) A graphene Nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis. Adv Funct Mater 20:453–459
Zhu Y, Murali S, Cai W, Li X, Suk JW, Potts JR, Ruoff RS (2010) Graphene and graphene oxide: Synthesis, properties, and applications. Adv Mater 22:3906–3924
Malard LM, Pimenta M, Dresselhaus G, Dresselhaus MS (2009) Raman spectroscopy in graphene. Phys Rep 473:51–87
Saeed AA, Sánchez JLA, O'Sullivan CK, Abbas MN (2017) DNA biosensors based on gold nanoparticles-modified graphene oxide for the detection of breast cancer biomarkers for early diagnosis. Bioelectrochem 118:91–99
Changfeng Z, Zhiyuan Z, Hai L, Fan L, Chunhai F, Hua Z (2013) Single-layer MoS2-based Nanoprobes for homogeneous detection of biomolecules. J Am Chem Soc 135:5998–6001
Yang R, Jin J, Chen Y, Shao N, Kang H, Xiao Z, Tang Z, Wu Y, Zhu Z, Tan W (2008) Carbon nanotube-quenched fluorescent oligonucleotides: Probes that fluoresce upon hybridization. J Am Chem Soc 130:8351–8358
Sebastian K, Swathi R (2008) Long range resonance energy transfer from a dye molecule to graphene has (distance)−4 dependence. J Chem Phys 120:086101
Lee C, Lin S, Wu Y, Hsieh Y (2018) Combining DNA-stabilized silver nanocluster synthesis with exonuclease III amplification allows label-free detection of coralyne. Anal Chim Acta 1042:86–92
Qian ZS, Shan XY, Chai LJ, Ma JJ, Chen JR, Feng H (2014) DNA nanosensor based on biocompatible graphene quantum dots and carbon nanotubes. Biosens Bioelectron 60:64–70
Yew YT, Loo AH, Sofer Z, Klímová K, Pumera M (2017) Coke-derived graphene quantum dots as fluorescence nanoquencher in DNA detection. Appl Mater Today 7:138–143
Bülbül G, Hayat A, Mustafa F, Andreescu S (2018) DNA assay based on nanoceria as fluorescence quenchers (nanoceracq DNA assay). Sci Rep 8:2426
Mu Q, Liu G, Yang D, Kou X, Cao N, Tang Y, Miao P (2018) Ultrasensitive detection of DNA based on exonuclease III-assisted recycling amplification and DNAzyme motor. Bioconjug Chem 29:3527–3531
Sun J, Guo A, Zhang Z, Guo L, Xie J (2011) A conjugated aptamer-gold nanoparticle fluorescent probe for highly sensitive detection of rHuEPO-α. Sensors 11:10490–10501
Heller MJ (2002) DNA microarray technology: Devices, systems, and applications. Annu Rev Biomed Eng 4:129–153
Acknowledgements
This work was partially supported by the Research Grants Council of the Hong Kong Special Administrative Region under Grant No. 16228216 and the National Natural Science Foundation of China under Grant No. 21705076. We would also like to acknowledge the supply of graphene oxide from World Linkage Holdings Limited (Hong Kong).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The author(s) declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 406 kb)
Rights and permissions
About this article
Cite this article
Iwe, I., Li, Z. & Huang, J. Graphene oxide and enzyme-assisted dual-cycling amplification method for sensitive fluorometric determination of DNA. Microchim Acta 186, 716 (2019). https://doi.org/10.1007/s00604-019-3825-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3825-7