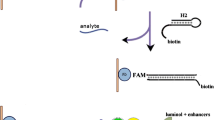
Abstract
An entropy-driven 3-D DNA walking machine is presented which involves catalytic hairpin assembly (CHA) for detection of microRNA. A 3-D DNA walking machine was designed that uses streptavidin-coated polystyrene microspheres as track carriers to obtain reproducibility. The method was applied to microRNA 21 as a model analyte. Continuous walking on the DNA tracks is achieved via entropy increase. This results in a disassembly of ternary DNA substrates on polystyrene microspheres and leads to cycling of microRNA 21. The release of massive auxiliary strands from ternary DNA substrates induces the CHA. This is accompanied by in increase in fluorescence, best measured at excitation/emission wavelengths of 480/520 nm. On account of entropy-driven reaction, the assay is remarkably selective. It can differentiate microRNA 21 from homologous microRNAs in giving a signal that is less than 5% of the signal for microRNA 21 except for microRNA-200b. The assay works in the 50 pM to 20 nM concentration range and has a 41 pM detection limit. The method displays good reproducibility (between 1.1 and 4.2%) and recovery (from 99.8 to 104.0%).

An entropy-driven 3-D DNA walking machine is described. It is based on the use of polystyrene microspheres and of a catalytic hairpin assembly reaction for sensitive microRNA detection. Figure Notes: AS represents auxiliary strand; S represents substrate strand; LS represents link strand; F represents fuel nucleic acid; RepF represents nucleic acid labeled with FAM; RepQ represents nucleic acid labeled with BHQ1.






Similar content being viewed by others
References
Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL (2000) A DNA-fuelled molecular machine made of DNA. Nature 406(6796):605–608
Zhuang JY, Lai WQ, Chen GN, Tang DP (2014) A rolling circle amplification-based DNA machine for microRNA screening coupling catalytic hairpin assembly with DNAzyme formation. Chem Commun 50(22):2935–2938
Hwang MT, Wang ZJ, Ping JL, Ban DK, Shiah ZC, Antonschmidt L, Lee J, Liu YS, Karkisaval AG, Johnson ATC, Fan CH, Glinsky G, Lal R (2018) DNA Nanotweezers and graphene transistor enable label-free genotyping. Adv Mater 30(34):1802440
Li LD, Li N, Fu SN, Deng YN, Yu CY, Su X (2019) Base excision repair-inspired DNA motor powered by intracellular apurinic/apyrimidinic endonuclease. Nanoscale 11(3):1343–1350
Lv SZ, Zhang KY, Zeng YY, Tang DP (2018) Double photosystems-based 'Z-Scheme' Photoelectrochemical sensing mode for ultrasensitive detection of disease biomarker accompanying three-dimensional DNA Walker. Anal Chem 90(11):7086–7093
AmicroRNA Y, Ben-Ishay E, Levner D, Ittah S, Abu-Horowitz A, Bachelet I (2014) Universal computing by DNA origami robots in a living animal. Nat Nanotechnol 9(5):353–357
Wilner OI, Willner I (2012) Functionalized DNA nanostructures. Chem Rev 112(4):2528–2556
Yuan L, Giovanni M, Xie JP, Fan CH, Leong DT (2014) Ultrasensitive IgG quantification using DNA nano-pyramids. Npg Asia Mater 6:e112
Hwang MT, Landon PB, Lee J, Choi D, Mo AH, Glinsky G, Lal R (2016) Highly specific SNP detection using 2D graphene electronics and DNA strand displacement. P Natl Acad Sci USA 113(26):7088–7093
Liu XQ, Lu CH, Willner I (2014) Switchable reconfiguration of nucleic acid nanostructures by stimuli-responsive DNA machines. Acc Chem Res 47(6):1673–1680
Li J, Fan CH, Pei H, Shi JY, Huang Q (2013) Smart drug delivery Nanocarriers with self-assembled DNA nanostructures. Adv Mater 25(32):4386–4396
Peng LC, Zhang P, Chai YQ, Yuan R (2017) Bi-directional DNA walking machine and its application in an enzyme-free Electrochemiluminescence biosensor for sensitive detection of MicroRNAs. Anal Chem 89(9):5036–5042
Liu MH, Cheng J, Tee SR, Sreelatha S, Loh IY, Wang ZS (2016) Biomimetic autonomous enzymatic Nanowalker of high fuel efficiency. ACS Nano 10(6):5882–5890
Wang DF, Vietz C, Schroder T, Acuna G, Lalkens B, Tinnefeld P (2017) A DNA Walker as a fluorescence signal amplifier. Nano Lett 17(9):5368–5374
Tomov TE, Tsukanov R, Glick Y, Berger Y, Liber M, Avrahami D, Gerber D, Nir E (2017) DNA bipedal motor achieves a large number of steps due to operation using microfluidics-based Interface. ACS Nano 11(4):4002–4008
Zhang P, Jiang J, Yuan R, Zhuo Y, Chai YQ (2018) Highly ordered and field-free 3D DNA nanostructure: the next generation of DNA Nanomachine for rapid single-step sensing. J Am Chem Soc 140(30):9361–9364
De Luna P, Mahshid SS, Das J, Luan BQ, Sargent EH, Kelley SO, Zhou RH (2017) High-curvature Nanostructuring enhances probe display for biomolecular detection. Nano Lett 17(2):1289–1295
Li W, Wang L, Jiang W (2017) A catalytic assembled enzyme-free three-dimensional DNA walker and its sensing application. Chem Commun 53(40):5527–5530
Zeng R, Luo Z, Su L, Zhang L, Tang D, Niessner R, Knopp D (2019) Palindromic molecular Beacon based Z-scheme BiOCl-au-CdS Photoelectrochemical biodetection. Anal Chem 91(3):2447–2454
Zeng RJ, Luo ZB, Zhang LJ, Tang DP (2018) Platinum Nanozyme-catalyzed gas generation for pressure-based bioassay using polyaniline nanowires-functionalized graphene oxide framework. Anal Chem 90(20):12299–12306
Yang XL, Tang YA, Mason SD, Chen JB, Li F (2016) Enzyme-powered three-dimensional DNA Nanomachine for DNA walking, payload release, and biosensing. ACS Nano 10(2):2324–2330
Qu XM, Zhu D, Yao GB, Su S, Chao J, Liu HJ, Zuo XL, Wang LH, Shi JY, Wang LH, Huang W, Pei H, Fan CH (2017) An exonuclease III-powered, on-particle stochastic DNA Walker. Angew Chem Int Ed 56(7):1855–1858
Wei W, Wei M, Yin LH, Pu YP, Liu SQ (2018) Improving the fluorometric determination of the cancer biomarker 8-hydroxy-2 '-deoxyguanosine by using a 3D DNA nanomachine. Microchim Acta 185(10):494
Yin D, Tao YY, Tang L, Li W, Zhang Z, Li JL, Xie GM (2017) Cascade toehold-mediated strand displacement along with non-enzymatic target recycling amplification for the electrochemical determination of the HIV-1 related gene. Microchim Acta 184(10):3721–3728
Liang CP, Ma PQ, Liu H, Guo XG, Yin BC, Ye BC (2017) Rational engineering of a dynamic, entropy-driven DNA Nanomachine for intracellular MicroRNA imaging. Angew Chem Int Ed 56(31):9077–9081
Zhang DY, Turberfield AJ, Yurke B, Winfree E (2007) Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318(5853):1121–1125
He L, Lu DQ, Liang H, Xie ST, Zhang XB, Liu OL, Yuan Q, Tan WH (2018) mRNA-initiated, three-dimensional DNA amplifier able to function inside living cells. J Am Chem Soc 140(1):258–263
Liu BW, Wu P, Huang ZC, Ma LZ, Liu JW (2018) Bromide as a robust Backfiller on gold for precise control of DNA conformation and high stability of spherical nucleic acids. J Am Chem Soc 140(13):4499–4502
Li YG, Cu YTH, Luo D (2005) Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nat Biotechnol 23(7):885–889
Peng W, Zhao Q, Chen M, Piao J, Gao W, Gong X, Chang J (2019) An innovative "unlocked mechanism" by a double key avenue for one-pot detection of microRNA-21 and microRNA-141. Theranostics 9(1):279–289
He MQ, Wang K, Wang WJ, Yu YL, Wang JH (2017) Smart DNA machine for carcinoembryonic antigen detection by exonuclease III-assisted target recycling and DNA Walker Cascade amplification. Anal Chem 89(17):9292–9298
Liu JT, Du P, Zhang J, Shen H, Lei JP (2018) Sensitive detection of intracellular microRNA based on a flowerlike vector with catalytic hairpin assembly. Chem Commun 54(20):2550–2553
Zhang Y, Luo SH, Bo ST, Chai ZX, Li B, Liu JM, Zheng L (2018) A novel electrochemical cytosensor for selective and highly sensitive detection of cancer cells using binding-induced dual catalytic hairpin assembly. Biosens Bioelectron 102:568–573
Yang WT, Zhou XX, Zhao JM, Xu WJ (2018) A cascade amplification strategy of catalytic hairpin assembly and hybridization chain reaction for the sensitive fluorescent assay of the model protein carcinoembryonic antigen. Microchim Acta 185(2):100
Bai SL, Wang T, Zhang Z, Sheng SC, Yu W, Xie GM (2017) A novel colorimetric biosensor for detecting target DNA and human alpha thrombin based on associative toehold activation concatemer induced catalyzed hairpin assembly amplification. Sensor Actuat B-Chem 239:447–454
Acknowledgments
This research work was financially supported by the National Natural Science Foundation of China (No. 81672112, 81702101), Chongqing Technology Innovation and Application Demonstration Project (cstc2018jscx-msybX0010) and Key Project of Education Department of Sichuan (No. 16ZA0181).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 7.11 mb)
Rights and permissions
About this article
Cite this article
Yang, T., Fang, J., Guo, Y. et al. Fluorometric determination of microRNA by using an entropy-driven three-dimensional DNA walking machine based on a catalytic hairpin assembly reaction on polystyrene microspheres. Microchim Acta 186, 574 (2019). https://doi.org/10.1007/s00604-019-3689-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3689-x