Abstract
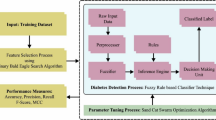
Healthcare organizations and Health Monitoring Systems generate large volumes of complex data, which offer the opportunity for innovative investigations in medical decision making. In this paper, we propose a beetle swarm optimization and adaptive neuro-fuzzy inference system (BSO-ANFIS) model for heart disease and multi-disease diagnosis. The main components of our analytics pipeline are the modified crow search algorithm, used for feature extraction, and an ANFIS classification model whose parameters are optimized by means of a BSO algorithm. The accuracy achieved in heart disease detection is \(99.1\%\) with \(99.37\%\) precision. In multi-disease classification, the accuracy achieved is \(96.08\%\) with \(98.63\%\) precision. The results from both tasks prove the comparative advantage of the proposed BSO-ANFIS algorithm over the competitor models.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Big Data analytics techniques help in improving the decision-making capability of any system characterized by “the five V” issues (variety, volume, veracity, velocity, validity, and volatility [40]). Healthcare is a area in which a large volume of data of the patients and other related data need to be analyzed and processed: often they are characterized also by issues related to variety, veracity and validity [54]. The main application of big data and machine learning in medicine is in the area of diagnostic, i.e., the detection/prediction of various diseases, most often formalized as a classification or regression problem. Big data bear the potential to contribute important results if combined with the methods of artificial intelligence, machine learning/data mining/computational intelligence/soft computing such as neural network and fuzzy logic.
The latter two families of methods are among the most significant in soft computing. Artificial neural networks (ANNs) correspond to one of the most prominent classes of inferential algorithms (i.e., algorithms that are able to learn models by generalizing the knowledge available in the training data). They are inspired by the structure and functionality of the brain. By composing elementary units, the artificial neurons, in various architectures, ANNs can learn very complex classification or regression models, typically by exploiting error back-propagation and gradient descent optimization techniques. They have been used for many years in a wide number of applications, and have gained further momentum recently, with the development of deep learning techniques [4, 5, 8, 12, 23, 44, 58]. Typically, ANNs are characterized by a black-box behavior: given the learned model, it is not easy to get an explanation about how the model gets its decisions [62].
Fuzzy set theory [67] represents an extension of the classical (often referred to as “crisp”) set theory; in fuzzy set theory, an element can belong to a set to some degree between zero and one (two ends included). Fuzzy logic is a many-valued extension of Boolean logic based on fuzzy set theory; the truth values of variables may be any real number in the unit interval. Fuzzy set theory and fuzzy logic were developed to model systems featuring characteristics such as imprecision, vagueness and graduality, and represent the ideal tools to handle perceptual data. Science, typically, tries to pass from perception to measurement, and then uses “crisp numbers” to feed “crisp models”; however, many situations occur in which there is the need to retain vagueness and imprecision into the reasoning; in those cases, fuzzy techniques can be very useful. This is especially true in the medical area, where often observations are not expressed by numbers, but rather by words (e.g., “severe cough,” “low mobility”) and where rules can also be expressed by fuzzy statements. Understandably, fuzzy logic has a wide range of applications in medicine [16, 53, 57, 64] and in area, such as engineering [6, 10, 11, 13, 14], image processing [22, 28, 31, 35, 59], as well as linguistics an other fields [30, 42]. A relevant class of applications is based fuzzy inference systems, which build input–output mappings based on fuzzy logic. However, those systems do not learn from examples, but apply rules created by domain experts to the input data; hence, the a-priori knowledge of domain experts is essential for building such systems. When that knowledge is available, they are of relatively simple interpretation and simple implementation (if their size is moderate).
A drawback of the standard artificial neural networks is that they can handle only crisp inputs, thus they would not be able to process many data taking the form of linguistic expressions; this is a shortcoming in the diagnosis of diseases [55]. This inability can be mitigated by the use of fuzzy logic if one is able to represent knowledge in the form of fuzzy sets. By contrast, fuzzy set theory and fuzzy logic are sometime cumbersome to use, if one needs to specify the rules by hand, but if one could learn at least part of the fuzzy rules from the data, the effort can be reduced considerably: suitable machine learning algorithms can be devised for this task.
The advantages and limitations of the two kinds of techniques have lead to the creation of a class of hybrid techniques called neuro-fuzzy systems, which try to exploit the advantages both of ANNs and of fuzzy logic. Their configurations are versatile, in that they can take either crisp or fuzzy inputs, learn fuzzy rules, and yield either fuzzy or crisp outputs. A specific form of neuro-fuzzy systems implementing a Sugeno-like fuzzy system (where the output memberships are linear function of the input values) was proposed by Jang in 1993 [29]; they are known as adaptive neural-fuzzy inference systems (or ANFIS). They use neural network learning methods (a mixture of gradient descent-based back-propagation and least mean squares steps) to tune the parameters of a fuzzy inference system. It is characterized by a wide choice of allowed membership functions, strong generalization abilities, good explanation capabilities and easiness in incorporating both linguistic and numeric knowledge [50]. Extended introductions neuro-fuzzy systems in general and to ANFIS in particular can be found in several texts, including [19] and [17].
ANFIS have been helpful in diseases diagnosis [52]. However, the applications of ANFIS are not bound to healthcare; they showed remarkable results in various fields of engineering, economics, transportation, etc. [65]. The major challenges in the use of the ANFIS can include their complexity, the higher computational cost, the complication of handling a large number of membership functions [51], and the difficulty in handling high-dimensional problems while working with big data or large datasets [38, 56]. Several of those issues were addressed by the researchers in the latest years, often with the help of nature-inspired evolutionary computation techniques.
In this paper, we presented a new combination of a beetle swarm optimization (BSO) algorithm with an adaptive neuro-fuzzy inference system—that we call BSO-ANFIS—for disease diagnosis and for the assessment of multi-disease in big healthcare data. In the BSO technique we choose, the beetle antennae search (BAS) meta-heuristic algorithm is combined with a particle swarm optimization (PSO) model. The BAS algorithm is inspired by the fact that beetles have long antennas, which help them to search food as well as to detect obstacles in their way. The main reason of using BAS is that it is less complex in comparison with many other existing optimization algorithms. Wang et al. [66] analyzed the detailed working of the BAS algorithm and found that it is highly sensitive to antennae positions at the initial stage for the iteration process; this yields poor performance for high-dimensional problems. To address this issue, the authors took the inspiration from the particle swarm optimization (PSO) algorithm, and came up with the new combination of BAS and PSO algorithm called BSO.
The proposed model is applied to heart disease classification and to multi-disease classification, based on a real-world dataset. The outline of the work we report about is the following.
-
In the preprocessing phase, we processed the multi-disease dataset using split and integration with the modified crow search algorithm (MCSA) for feature extraction.
-
The ANFIS model was applied for the assessment of various types of diseases. The parameters of ANFIS were optimized using the beetle swarm optimization (BSO) algorithm.
-
The proposed BSO-ANFIS model’s performance was compared to existing models for the big healthcare data analysis for multi-diseases and heart health.
The BSO-ANFIS model was implemented in MATLAB using the available libraries.
The reminder of the paper is structured as follows. The related work of healthcare data analysis is presented in Sect. 2. The proposed BSO-ANFIS model for multi-disease analysis is discussed in Sect. 3. Section 4 reports the performance of the proposed BSO-ANFIS compared to other existing models. Section 5 reports our conclusions.
2 Related work
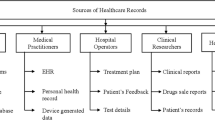
Healthcare organizations, government agencies, and research in the pharmaceutical industry produce a large amount of data from patient care, and from record keeping. The data are generated through the most disparate sources, such as wearable devices, medical imaging, payee records, and electronic health records. So there is a lot of variability in the context, type, and format of the data. This large amount of data requires storage and retrieval services, to support health management, decision support systems in clinics, and regular surveillance of diseases. These opportunities raise the requirements for big healthcare data knowledge discovery for accountable, result-oriented, patient-centric, and evidence-based care. Healthcare data are affected also by several issues related to privacy protection; thus, the data analytic platforms are required to safeguard the privacy of the patients. For overviews of this area of research, one can see Sughasiny et al. [63] and Alexandra et al. [41].
Nilashi et al. [48] proposed a method for the diagnosis of heart diseases using principal component analysis and support vector machines (SVMs) based on fuzzy logic (FSVM). It also used special techniques for the imputation of missing values. The method was based on incremental learning. However, the work used only the two real-world datasets (named Cleveland and Statlog) that were not very large, a big limitation for an approach that aims at working with real-world clinical datasets.
Gougam et al. [24] used a neuro-fuzzy inference system for the diagnosis of faults in a bearing systems. The input of the diagnosis was the data coming from a signal processing technique known as autogram analysis. The inference system could predict the remainder lifetime time of the element hampering the mechanism. This helps increasing the lifespan of the bearing. As the fuzzy inference system can predict the problems in bearings using the input data, in the same way, it can be accurately used for the detection of various diseases.
Mohanty et al. [46] gave a clear description of how machine learning technologies, including the different kinds of artificial neural networks can help in the detection of the various diseases. These techniques can work both in biomedical as well as healthcare domains. The evolution of such techniques can help the patients in the prediction of various diseases, and also used for self-diagnosis. There are a number of techniques that are given for disease detection which includes Bayesian networks, wavelet networks, and Gaussian mixture models. In this paper, the author tries to highlight the advantages of the use of ANFIS in the healthcare sector.
Hu et al. [27] proposed a diagnosis model for doctors, named simultaneously aided diagnosis model (SADM), which proved to be helpful in improving the detection performance. The model was compared with support vector machines (SVM) and neural networks (ANN). SVM and ANN were earlier used with datasets of hyperlipemia disease, and the SADM model proved to be more accurate. The only limitation for this algorithm was that the accuracy turned out to be very sensitive to the details of the preprocessing and feature selection phases.
Manogaran et al. [43] suggested a method based on deep learning for the diagnosis of heart diseases: multiple kernel learning with adaptive neuro-fuzzy inference system (MKL with ANFIS). Then, the inference system was used as a classifier and compared to other ML tchniques such as least square with support vector machine (LS-SVM), general discriminant analysis and LS-SVM, principal component analysis with ANFIS and latent dirichlet allocation with ANFIS. However, the experimentation was limited to a single datatset: the KEGG metabolic reaction network dataset.
Nair et al. [47], using apache spark, developed a cloud-based model for handling data related to healthcare. It consisted of a remote-access system that helped in predicting the status of patients; it suggested the necessary actions and precautions be taken by the patients after predicting their diseases using different machine learning models, including decision trees. The system proved helpful in a number of cases. The system can be linked with various providers of healthcare for helping the patients in deciding whether they need to meet doctor for further assistance.
Aujla et al. [9] used the software defined network (SDN) in multiedge-cloud environment for the management of coarse grained application such as healthcare, e-commerce, and banking. The flow of network scheduled energy efficiently using a control scheme. The experimental evaluation was carried on using traces from the Google workload. The proposed scheme showed the effectiveness in big data processing for optimal decision making.
Chaudhary et al. [15] developed a big data management technique based on SDN for the optimization of storage and network bandwidth. The rule-and-action-based technique employed bloom-filters in the OpenFlow controller. Big data applications can be deployed and analyzed in real time with the help of this scheme.
Data leakage is one of the most relevant issues in internet-based healthcare applications. Aujla et al. [7] controlled the deduplication of big data in the cloud environment using a Merkle hash-tree, whereas Singh et al. [61] proposed a intrusion detection system using dew computing as a service. Kaur et al. [34] provided a solution for the problem of dimensionality reduction for big data on the smart grid. Similar solutions could be applied to healthcare data generated from various IoT devices.
Ed-daoudy [18] presented a method that can predict breast cancer using different machine learning techniques, including decision trees. This offline system used apache spark and proved to be very effective for the prediction of health diseases in large data sets. It proved to be qualitatively and quantitatively better than the traditional systems in terms of complexity, processing time as well as development time. This also implies the additional advantage of saving time and money in prediction.
Ahmed et al. [1] presented a machine learning technique that can predict heart diseases based on the healthcare data of the patients. This paper guides in the choice of the appropriate machine learning algorithm from the set of algorithms given by decision trees, random forest, SVM, and various other classifiers. The accuracy was enhanced using various tuning and cross-validation parameters. This prediction system was developed using Apache Spark and Apache Kafka.
Silahtarouglu et al. [60] implemented two different models for predicting various diseases in the big data sets of healthcare data; a neural network-based technique and and a random forest-based technique. It could perform diagnoses based on complaints given verbally to the system by the patients and could find a natural application in supporting patients that contact healthcare centers. A drawback of this system is that it needs to be implemented with utmost care, furthermore the diagnosis by the doctor can not be compared during emergency conditions.
Khan et al. [37] developed a framework, using multilayer perceptrons and spark-based models, that can analyze the healthcare data from heterogeneous sources. This technique was used both for binary and multi-class classification problems; the accuracy was higher in comparison with the traditional techniques, but it required a considerable amount of preprocessing.
3 The proposed multi-disease analysis model
3.1 Data preprocessing
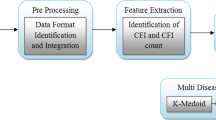
Data preprocessing is required once we get the electronic healthcare records (EHR) from the various sources. The first step is data cleaning, to handle the missing and spurious values and perform the normalization [20]. The featured data may contain unwanted spaces, symbols, or words. Those elements are removed by using a stop-word elimination techniques. The noise from the data set must be removed to help improving the accuracy of the analytic models.
The data available related to healthcare is mostly unstructured. It is hard to aggregate or integrate the healthcare data due to their diverse nature. We integrated the data by matching and unifying their formats. Eventually, we obtained a number of comma-separated values (CSV) text files, each with a maximum size of 1 GB. To handle the missing values in the dataset, we replaced them with the median of the available values for the same variable.
3.2 Datasets and feature selection
3.2.1 First experiment, used features
For the first experiment, we used the dataset Framingham and the dataset Hungarian, collected from the Kaggle repository [33]. Out of the 76 available attributes, we keep the same 13 selected by Khan and Algarni [36]: Age (Young, Medioum, Old, Very Old), Sex (Male, Female), Chest pain (Typical Angina, Atypical Angina, Non-Anginal pain, Asymptomatic), Resting blood pressure (Low, Medium, High, Very high), Serum cholesterol (Low, Medium, High, Very high), Fasting blood sugar (> \(120\,{\mathrm{mg}}/{\mathrm{dl}}\) Yes/No), Resting electrocardiographic results (Normal,ST-T wave abnormality, Left ventricular hypertrophy), Maximum heart rate (Low, Medium, High, Very high), Exercise-induced angina (Yes/No), Depression induced by exercise relative to test (Low, Risk, Terrible), Slope of the peak exercises ST segment (Up-sloping, Flat, Down-sloping), Major vessels (0–3), Thallium scan (Normal, Fixed defect, Reversible defect)). The target variable for this dataset was a binary variable (Disease: Yes/No).
3.2.2 Second experiment, feature selection
The real-world dataset we used for the second experiment was the Health Dataset from the official USA Health site [26] covering the data from ten counties (Cleburne, Clay, Chilton, Calhoun, Butler, Bullock, Blount, Barbour, Baldwin, Autauga). The dataset contains a very large number of features. A large number of techniques is available for feature selection [21, 39]. We selected the features by applying a nature-inspired evolutionary optimization algorithm, the modified crow search algorithm (MCSA) developed by Gupta et al. [25]. In MCSA, each crow searches the environment to find food and saves it in hiding places; each crow also stealthily follows other crows to find out their hiding places. Each crow memorizes the best solution found so far (each crow has a memory, hereafter denoted by mem, to store the hiding places): the goodness of the solution is evaluated by a fitness function. CSA has only two controlling parameters: flight length (hereafter denoted by fl) and awareness probability (the awareness of a crow that he is followed by another crow, hereafter denoted by Ap). MCSA adds a refined destination selection mechanism and a mechanism for adapting the flight length. The position of the i-th crow hereafter is denoted by \(p_i\) and represents a feasible solution in the solution space. For the use of MCSA in feature selection the solution space all the possible combination of features. To this purpose, the solution space can be represented as a d dimensional cube: any point within the volume of that cube can be mapped to a feasible solution by rounding each dimension to the closest integer (1 will mean inclusion, 0 exclusion) before feeding a candidate solution to a fitness function. The fitness function for a feature selection problem can be any base classifier, typically a low cost classifier, such as the k-nearest-neighbor classifier The following steps are involved in feature selection using the MCSA approach.
-
1.
We consider there are n number of big data files with for healthcare records. The first step consists of splitting the data attribute wise into small portions. Each split part is considered as a crow \(BD_i = \{c_1, c_2, c_3, \ldots , c_n\}\). Random numbers representing initial positions in the search space are assigned to each crow.
-
2.
Two possible situations can occur: the crow can be aware or unaware of the fact of being followed. The awareness of a crow is decided by generating a random number \(a_i\in [0,1]\) and checking whether it passes the awareness probability threshold Ap. When the crow does not know that he is being followed, the position is updated based on Eq. 1, where \(rv_i\in [0,1]\) is a random number, and t denotes the time-step.
$$\begin{aligned} p_{i,t+1}~ =~ p_{i,t}~ +~ rv_{i}~ \times ~ fl ~\times ~ (mem_{i,t} ~-~ p_{i,t}) \quad { if }\quad a_i>Ap \end{aligned}$$(1)otherwise, if the crow is aware of being followed, the new position is generated at random.
-
3.
Further, the memory containing the position of the crow is updated. The fitness function calculates the fitness value of the new position of the crow. If the new fitness value is larger then the current fitness value, update the memory.
-
4.
Repeat the entire process till a maximum number of iterations is reached or a sufficiently stable optimum is found.
The main features eventually considered in our study were Year, Age, Country name, Death ratio, Symptoms, and Treatment. The target variable was disease name, which could take the following values: Alzheimer, Anemia, Asthma, Diabetes, Fibroid tumor, Gastroenteritis, Heart disease, Hemorrhoids, Hepatitis, Kidney disease, Neuropathy, Parkinson Disease. Notably, Symptoms and Treatment are linguistic variables. The number of symptoms for each disease is shown in Fig. 1. The treatment was indicating only the kind of prescription (ointment, spray, suspension, lotion, tablets, injections, or powder).
3.3 Proposed BSO-ANFIS model
In this section, we discuss the proposed BSO-ANFIS model for multi-disease classification. Jang [29] presented the ANFIS model with the nonlinear mapping of Takagi-Sugeno inference using IF-THEN fuzzy rules. The model is structured in five layers, illustrated in Fig. 2. The performance of ANFIS depends upon the design of rules. The number of rules depends upon the number of input and tunable parameters. The fundamental rules of ANFIS is defined in Eqs. 2 and 3.
-
Rule 1: If \(FV_{1}\) is \(A_{i}\) and \(FV_{2}\) is \(B_{i}\) then
$$\begin{aligned} RL_{i} = s_{i}.FV_{i} + t_{i}.FV_{i+1} + u_i \end{aligned}$$(2) -
Rule 2: If \(FV_{1}\) is \(A_{i+1}\) and \(FV_{2}\) is \(B_{i+1}\) then
$$\begin{aligned} RL_{i+1} = s_{i+1}.FV_{i} + t_{i+1}.FV_{i+1} + u_{i+1} \end{aligned}$$(3)
Here, the fuzzy sets are denoted by \(A_{i}\), \(A_{i+1}\), \(B_{i}\),\(B_{i+1}\). The training process required the parameters represented by \(u_{i}\), \(u_{i+1}\), \(t_{i}\), \(t_{i+1}\), \(s_{i}\), \(s_{i+1}\). The BSO is applied on these parameters to optimize the performance of ANFIS. The detailed working of BSO for optimizing the parameter explain in the next section. The description of ANFIS follows:
-
Layer 1: The first layer is known as the fuzzification layer. The purpose of fuzzification is to determine the membership function and input values. The nodes in the fuzzification layer are adaptive nodes. Equations 4 and 5 describe the node function.
$$\begin{aligned} NF_{1,i}=\, & {} \mu _{A_i} (a) \end{aligned}$$(4)$$\begin{aligned} NF_{1,i}=\, & {} \mu _{B_i} (b) \end{aligned}$$(5)The inputs are denoted by a and b. The fuzzy sets \(A_i\) and \(B_i\) utilize the degree of membership of a and b represented by \(\mu _{A_i}\) and \(\mu _{B_i}\) respectively.
-
Layer 2: The responsibility of the second layer is to take the input signal, find the product, and give the output as per Eq. 6. The strength of the rules is generated in this layer.
$$\begin{aligned} NF_{2,i} = P_{i} = \mu _{A_i}(a) * \mu _{B_i}(b) \end{aligned}$$(6) -
Layer 3: The firing strength is normalized in this layer. The normalization \(\hat{P_{i}}\) is calculated as per Eq. 7 for node i.
$$\begin{aligned} NF_{3,i} = \hat{P_{i}} = \frac{P_{i}}{P_1 + P_2} \quad where \quad i= 1,2 \end{aligned}$$(7) -
Layer 4: In the fourth layer, the node function of adaptive nodes are expressed in Eq. 8.
$$\begin{aligned} NF_{4,i} = \hat{P_{i}}f_i = \hat{P_{i}}(p_{i}a ~+~ q_{i}b ~+~ r_{i}) \end{aligned}$$(8)The consequent parameters defined as \(p_i, q_i, r_i\) are normalized values. Also, inputs a, b are used here.
-
Layer 5: The final output is calculated in this layer as per Eq. 9. It is the addition of layer 4 output.
$$\begin{aligned} NF_{5} = \sum _{i} \hat{P_i} f_{i} = \frac{\sum _{i}~P_{i} f_{i}}{\sum _{i} P_{i}} \end{aligned}$$(9)
3.4 Optimization of parameters using BSO
The ANFIS parameters are optimized using a beetle swarm optimization (BSO) technique. The BSO optimization algorithm we chose is the combination of particle swarm optimization (PSO) and of an algorithm inspired by the foraging nature of beetles. This derives from the observation that the nearby environment is analyzed by beetles through their antennae. The beetle moves in that direction of the antennae where the food smells are most concentrated. a meta-heuristic optimization algorithm as per the behavior of beetle was designed by Jiang et al. [32]. The steps involved in the optimization of ANFIS using BSO are shown in Fig. 3. The kernel-based clustering approach was applied to partition the dataset into various clusters. Furthermore, the fuzzy rules for ANFIS were initialized using the cluster centers. After that, the ANFIS model was trained with the help of BSO model.
The position of the beetle was determined in the space of Q-dimensional at i as \(q^{t}\). The position of beetle has been determined using Eq. 10.
with
The beetle searching direction is chosen uniformly at random and is denoted by \({\hat{d}}\). The length of the beetle searching step is denoted with \(s^{i}\), whereas \(d^{i}\) is the distance that can be senses by the antennae. These parameters are set to a high value at the beginning and decrease over time; thus one first tries to access a wide region, then one reduces so as to obtain a reasonable capacity for the beetle. The right detected position is expressed with \(q_{ri}\), and left position detected is denoted with \(q_{li}\). These positions are equipped with food flavor, the food concentration is denoted the fitness function values \(f(q_{ri})\) and \(f(q_{li})\) calculated from the proposed algorithm; symFun is the function of the symbol.
The position of beetle are affected from the update of speed and information gathered through the antennae. Consider the beetle swarm as \(B~={B_1,~B_2,~\ldots ,~B_n}\) with n size of population. Searching space is in D dimensions. The position of beetle i in the D dimensional searching space is represented with \(B_i = (b_{i1},~b_{i2},~\ldots ~,~b_{iD})^T\) with solution of optimization. Beetle speed is described as \(S_i = (s_{i1},~s_{i2},~\ldots ~,~s_{iD})^T\). As beetle are approaching to the extreme global value, the speed of every beetle varies accordingly. Further, the extreme of individual beetle is denoted with \(E_i = (e_{i1},~e_{i2},~\ldots ~,~e_{iD})^T\). The global extreme is described with \(E_g = (e_{g1},~e_{g2},~\ldots ~,~e_{gD})^T\). The process of BSO algorithm for position and speed update is as follows:
The value of \(i={1,2,\ldots ,n}\), \(d={1,2,\ldots ,D}\), and j is for each iteration. The value of displacement is represented with \(\mu\), which is obtained form the antennae of beetle. The symbols \(\alpha\) and \(\theta\) in Eqs. 13 and 14 are denoting the loosening factor and inertia weight, respectively. We can also adjust these parameters. The impact degree is defined with \(m_1\) and \(m_2\), whereas \(r_1\) and \(r_2\) depicts the random functions.
First, we performed the environmental modeling. The target points were selected for the input in the given environment. The parameters of BSO algorithms such as \(\alpha\), \(\theta\), \(m_1\), and \(m_2\) were initialized. The speed \(S_i\) and position \(B_i\) was also initialized randomly in the model. Then, the fitness function of the each beetle was calculated and the value in extreme individual of beetle was put in \(E_i\). The minimum value was found from the \(E_i\) extreme individual value and the extreme global \(E_g\) declared. After that, we updated the beetle swarm attributes with the help of Eqs. 13, 16 and 17. The extreme value \(E_i\) and global extreme values \(E_g\) were updated by calculating the fitness function. The value \(h^{t}\) was initialized with \(h^{t} = 0.01 + 0.95 h^{t-1}\), set high at the start, was then decreased with each iteration, denoted here with \({\hat{h}}\). The iterations were then performed up to optimization.
The global extreme values \(E_g\) obtained by this algorithm are considered to be optimal parameters for the ANFIS model.
In both experiments, the simulation of the proposed model was performed on MATLAB using the built-in libraries of ANFIS. The fuzzy logic toolbox software in MATLAB provides a command-line function (anfis) and an interactive app (neuro-fuzzy designer) for training an adaptive neuro-fuzzy inference system (ANFIS). Using ANFIS training methods, one can design, train, and test Sugeno-type fuzzy inference systems.
4 Performance evaluation
In the first experiment, the performance evaluation of BSO-ANFIS has been performed using standard metrics for binary classification: Accuracy, Precision (a.k.a. True Positive Rate), Sensitivity (a.k.a. Recall), and Specificity (True Negative Rate). We recall hereafter their definitions for the sake of convenience. Let TP denote the number of True Positives, TN the number of True Negatives, FP the number of False Positives and FN the number of False Negatives, then \(Accuracy= (TP+TN)/(TP+FP+TN+FN)\), \(Precision = TP/(FP+TP)\), \(Sensitivity = TP/(TP+FN)\), \(Specificity = TN/(TN+FP)\). The confusion matrix in Fig. 4 illustrates the color codes used in the following figures for the different quantities just defined (including the Negative Prediction Value, NPV, not reported in Table 1).
4.1 First experiment: heart disease detection
In this experiment, we compare the performance of BSO-ANFIS to that of ANFIS, as well to that of other published results. We broke the whole dataset in training, validation and test set and used tenfold cross-validation. The test set performance is shown in Fig. 5. One can see that the performance of the proposed BSO-ANFIS is extremely good as compared to ANFIS. For instance in terms of accuracy, ANFIS correctly classified 2720 records out of 3200, whereas the proposed BSO-ANFIS is able to classify 3180 out of 3200 records. As to sensitivity we have that out of 1800 patients, ANFIS identifies 1420, whereas the BSO-ANFIS identifies 1775.
In Table 1, we compare the test accuracy, precision, sensitivity and specificity of BSO-ANFIS to those of other algorithms: HODBNN [2], HRFLM [45], \(\chi ^{2}-DNN\) [3] and ICA(MH) [49]. The results demonstrate that the proposed BSO-ANFIS model globally outperforms the other models.
4.2 Second experiment: multi-disease identification
The performance of proposed BSO-ANFIS and other models has been evaluated using accuracy and precision. The other algorithms considered were artificial neural networks (ANN), support vector machines (SVM), conventional ANFIS. Also here, we broke the whole dataset in training, validation and test set and used tenfold cross-validation. The results are reported in Table 2 and Fig. 6. The overall accuracy of the proposed BSO-ANFIS model is \(96.08\%\), which is higher as compare to ANN, SVM, and conventional ANFIS model. The accuracy of the models are: ANN (\(93.42\%\)), SVM (\(91.6\%\)), ANFIS (\(93.58\%\)). The precision of BSO-ANFIS for multi-disease analysis is \(98.63\%\). The precision of the other models are: ANN (\(95.58\%\)), SVM (\(96.02\%\)), conventional ANFIS (\(93.58\%\)). The precision of the Neural Network is lower among the existing techniques. This proves the superiority of the BSO-ANFIS approach with respect to the other techniques.
5 Conclusions
In this paper, we proposed a model combining beetle swarm optimization and an adaptive neuro-fuzzy inference system for heart disease and multi-disease big healthcare data analysis. The feature selection for the second experiment is performed using a modified version of the crow search algorithm (CSA). The dataset is first to split and further integrated for the proper feature extraction. The heart disease dataset was obtained from Kaggle. The multi-disease dataset was obtained from the USA healthcare and services department. In the first case study, the performance of the proposed BSO-ANFIS model is compared with HODBNN, HRFLM, \(\chi ^{2}-DNN\), and ICA(MH) model for heart disease analysis. In the second case study, the performance of multi-disease analysis is obtained for a neural network, a support vector machine, and a conventional ANFIS. Using our model, we obtained \(99.1\%\) accuracy with \(99.37\%\) precision for heart disease classification and \(96.08\%\) accuracy with \(98.63\%\) precision in multi-disease classification. Both case studies demonstrate the superiority of the proposed BSO-ANFIS model over the existing models.
In the future, the work will be carried out to integrate the proposed model in the healthcare monitoring system. The accuracy of the proposed BSO-ANFIS model can be further investigated for cardiovascular diseases. The images, audio, or video data can be integrated to increase the accuracy of the decision support system. Furthermore, we plan to test the classification models in other application domains.
References
Ahmed H, Younis EM, Hendawi A, Ali AA (2020) Heart disease identification from patients’ social posts, machine learning solution on spark. Fut Gen Comput Syst 111:714–722
Al-Makhadmeh Z, Tolba A (2019) Utilizing iot wearable medical device for heart disease prediction using higher order boltzmann model: a classification approach. Measurement 147:106815
Ali L, Rahman A, Khan A, Zhou M, Javeed A, Khan JA (2019) An automated diagnostic system for heart disease prediction based on \({\chi ^{2}}\) statistical model and optimally configured deep neural network. IEEE Access 7:34938–34945
Almazrouei E, Gianini G, Almoosa N, Damiani E (2019) A deep learning approach to radio signal denoising. In: 2019 IEEE Wireless Communications and Networking Conference Workshop (WCNCW), pp 1–8. IEEE
Almazrouei E, Gianini G, Mio C, Almoosa N, Damiani E (2019) Using autoencoders for radio signal denoising. In: Proceedings of the 15th ACM International Symposium on QoS and Security for Wireless and Mobile Networks, pp 11–17
Ardagna CA, Bellandi V, Bezzi M, Ceravolo P, Damiani E, Hebert C (2018) Model-based big data analytics-as-a-service: take big data to the next level. IEEE Trans Serv Comput
Aujla GS, Chaudhary R, Kumar N, Das AK, Rodrigues JJ (2018) Secsva: secure storage, verification, and auditing of big data in the cloud environment. IEEE Commun Mag 56(1):78–85
Aujla GS, Jindal A, Chaudhary R, Kumar N, Vashist S, Sharma N, Obaidat MS (2019) Dlrs: deep learning-based recommender system for smart healthcare ecosystem. In: ICC 2019-2019 IEEE International Conference on Communications (ICC), pp 1–6. IEEE
Aujla GS, Kumar N, Zomaya AY, Ranjan R (2017) Optimal decision making for big data processing at edge-cloud environment: an sdn perspective. IEEE Trans Ind Inform 14(2):778–789
Bellandi V, Cimato S, Damiani E, Gianini G (2016) Possibilistic assessment of process-related disclosure risks on the cloud. In: Computational Intelligence and Quantitative Software Engineering, pp 173–207. Springer
Bellini E, Ceravolo P, Nesi P (2017) Quantify resilience enhancement of uts through exploiting connected community and internet of everything emerging technologies. ACM Trans Internet Technol 18(1):1–34
Buffoni F, Gianini G, Damiani E, Granitzer M (2018) All-implicants neural networks for efficient boolean function representation. In: 2018 IEEE International Conference on Cognitive Computing (ICCC), pp 82–86. IEEE
Ceravolo P, Bellini E (2019) Towards configurable composite data quality assessment. In: 2019 IEEE 21st Conference on Business Informatics (CBI), vol 1, pp 249–257. IEEE
Ceravolo P, Damiani E, Viviani M (2006) Bottom-up extraction and trust-based refinement of ontology metadata. IEEE Trans Knowl Data Eng 19(2):149–163
Chaudhary R, Aujla GS, Kumar N, Rodrigues JJ (2018) Optimized big data management across multi-cloud data centers: software-defined-network-based analysis. IEEE Commun Mag 56(2):118–126
Chen L, Yang X, Jeon G, Anisetti M, Liu K (2020) A trusted medical image super-resolution method based on feedback adaptive weighted dense network. Artif Intell Med 101857
Czogala E, Leski J (2012) Fuzzy and neuro-fuzzy intelligent systems. Physica 47
Ed-daoudy A, Maalmi K (2018) Application of machine learning model on streaming health data event in real-time to predict health status using spark. In: 2018 International Symposium on Advanced Electrical and Communication Technologies (ISAECT), pp 1–4. IEEE
Fullér R (2000) Introduction to neuro-fuzzy systems, vol 2. Springer, Berlin
Gianini G, Damiani E (2004) An ontology-driven approach to metadata design in the mining of software process events. In: International Conference on Knowledge-Based and Intelligent Information and Engineering Systems, pp 321–327. Springer
Gianini G, Fossi LG, Mio C, Caelen O, Brunie L, Damiani E (2020) Managing a pool of rules for credit card fraud detection by a game theory based approach. Fut Gen Comput Syst 102:549–561
Gianini G, Rizzi A (2017) A fuzzy set approach to retinex spray sampling. Multimed Tools Appl 76(23):24723–24748
Goodfellow I, Bengio Y, Courville A, Bengio Y (2016) Deep learning, vol 1. MIT press Cambridge, Cambridge
Gougam F, Rahmoune C, Benazzouz D, Varnier C, Nicod J (2020) Health monitoring approach of bearing: application of adaptive neuro fuzzy inference system (anfis) for rul-estimation and autogram analysis for fault-localization. In: 2020 Prognostics and Health Management Conference (PHM-Besançon), pp 200–206. IEEE
Gupta D, Rodrigues JJ, Sundaram S, Khanna A, Korotaev V, de Albuquerque VHC (2018) Usability feature extraction using modified crow search algorithm: a novel approach. Neural Comput Appl 1–11
HealthData: U.S. department of health and human services (accessed: 01.10.2020). https://healthdata.gov/
Hu Y, Duan K, Zhang Y, Hossain MS, Rahman SMM, Alelaiwi A (2018) Simultaneously aided diagnosis model for outpatient departments via healthcare big data analytics. Multimed Tools Appl 77(3):3729–3743
Ikegami Y, Sakurai Y, Sakai M, Fujikawa H, Tsuruta S, Gonzalez A, Sakurai E, Damiani E, Kutics A, Knauf R (2018) A visual counseling agent avatar with voice conversation and fuzzy response. In: 2018 World Automation Congress (WAC), pp 1–5. IEEE
Jang JS (1993) Anfis: adaptive-network-based fuzzy inference system. IEEE Trans Syst Man Cybern 23(3):665–685
Jeon G, Anisetti M, Damiani E, Monga O (2018) Real-time image processing systems using fuzzy and rough sets techniques
Jeon G, Anisetti M, Wang L, Damiani E (2016) Locally estimated heterogeneity property and its fuzzy filter application for deinterlacing. Inf Sci 354:112–130
Jiang X, Li S (2017) Bas: beetle antennae search algorithm for optimization problems. arxiv. arXiv preprint arXiv:1710.10724
Kaggle: Kaggle open dataset. last accessed: Dec. 31, 2020. [online], available: https://www.kaggle.com/datasets
Kaur D, Aujla GS, Kumar N, Zomaya AY, Perera C, Ranjan R (2018) Tensor-based big data management scheme for dimensionality reduction problem in smart grid systems: Sdn perspective. IEEE Trans Knowl Data Eng 30(10):1985–1998
Kerre EE, Nachtegael M (2013) Fuzzy techniques in image processing. Physica 52
Khan MA, Algarni F (2020) A healthcare monitoring system for the diagnosis of heart disease in the iomt cloud environment using msso-anfis. IEEE Access 8:122259–122269
Khan MA, Karim M, Kim Y et al (2018) A two-stage big data analytics framework with real world applications using spark machine learning and long short-term memory network. Symmetry 10(10):485
Krizea M, Gialelis J, Koubias S (2019) Comparative study between fuzzy inference system, adaptive neuro-fuzzy inference system and neural network for healthcare monitoring. In: 2019 8th Mediterranean Conference on Embedded Computing (MECO), pp 1–4. IEEE
Legesse M, Gianini G, Teferi D (2016) Selecting feature-words in tag sense disambiguation based on their shapley value. In: 2016 12th International Conference on Signal-Image Technology & Internet-Based Systems (SITIS), pp 236–240. IEEE
Li Y, Yu M, Xu M, Yang J, Sha D, Liu Q, Yang C (2020) Big data and cloud computing. Manual of Digital Earth p 325
L’heureux A, Grolinger K, Elyamany HF, Capretz MA, (2017) Machine learning with big data: challenges and approaches. IEEE Access 5:7776–7797
Mamdani EH (1977) Application of fuzzy logic to approximate reasoning using linguistic synthesis. IEEE Trans Comput 12:1182–1191
Manogaran G, Varatharajan R, Priyan M (2018) Hybrid recommendation system for heart disease diagnosis based on multiple kernel learning with adaptive neuro-fuzzy inference system. Multimed Tools Appl 77(4):4379–4399
Mio C, Gianini G (2019) Signal reconstruction by means of embedding, clustering and autoencoder ensembles. In: 2019 IEEE Symposium on Computers and Communications (ISCC), pp 1–6. IEEE
Mohan S, Thirumalai C, Srivastava G (2019) Effective heart disease prediction using hybrid machine learning techniques. IEEE Access 7:81542–81554
Mohanty R, Solanki SS, Mallick PK, Pani SK A classification model based on an adaptive neuro-fuzzy inference system for disease prediction. In: Bio-inspired Neurocomputing, pp 131–149. Springer
Nair LR, Shetty SD, Shetty SD (2018) Applying spark based machine learning model on streaming big data for health status prediction. Comput Electr Eng 65:393–399
Nilashi M, Ahmadi H, Manaf AA, Rashid TA, Samad S, Shahmoradi L, Aljojo N, Akbari E (2020) Coronary heart disease diagnosis through self-organizing map and fuzzy support vector machine with incremental updates. Int J Fuzzy Syst pp 1–13
Nourmohammadi-Khiarak J, Feizi-Derakhshi MR, Behrouzi K, Mazaheri S, Zamani-Harghalani Y, Tayebi RM (2019) New hybrid method for heart disease diagnosis utilizing optimization algorithm in feature selection. Health Technol pp 1–12
Panella M (2012) A hierarchical procedure for the synthesis of anfis networks. Adv Fuzzy Syst 2012
Salleh MNM, Talpur N, Hussain K (2017) Adaptive neuro-fuzzy inference system: Overview, strengths, limitations, and solutions. In: International Conference on Data Mining and Big Data, pp 527–535. Springer
Sarangi L, Mohanty MN, Patnaik S (2016) Design of anfis based e-health care system for cardio vascular disease detection. In: International Conference on Intelligent and Interactive Systems and Applications, pp 445–453. Springer
Schuh C (2005) Fuzzy sets and their application in medicine. In: NAFIPS 2005-2005 Annual Meeting of the North American Fuzzy Information Processing Society, pp 86–91. IEEE
Shafqat S, Kishwer S, Rasool RU, Qadir J, Amjad T, Ahmad HF (2020) Big data analytics enhanced healthcare systems: a review. J Supercomput 76(3):1754–1799
Shahid N, Rappon T, Berta W (2019) Applications of artificial neural networks in health care organizational decision-making: a scoping review. PloS One 14(2):e0212356
Sharma D, Singh Aujla G, Bajaj R (2019) Evolution from ancient medication to human-centered healthcare 4.0: a review on health care recommender systems. Int J Commun Syst e4058
Sharma D, Singh Aujla G, Bajaj R (2020) Deep neuro-fuzzy approach for risk and severity prediction using recommendation systems in connected health care. Trans Emerg Telecommun Technol e4159
Sharma S, Dudeja RK, Aujla GS, Bali RS, Kumar N (2020) Detras: deep learning-based healthcare framework for iot-based assistance of alzheimer patients. Neural Comput Appl 1–13
Shi J, Wu J, Anisetti M, Damiani E, Jeon G (2017) An interval type-2 fuzzy active contour model for auroral oval segmentation. Soft Comput 21(9):2325–2345
Silahtaroğlu G, Yılmaztürk N (2019) Data analysis in health and big data: a machine learning medical diagnosis model based on patients’ complaints. Commun Stat-Theory Methods pp 1–10
Singh P, Kaur A, Aujla GS, Batth RS, Kanhere S (2020) Daas: Dew computing as a service for intelligent intrusion detection in edge-of-things ecosystem. IEEE Internet of Things Journal
Stier J, Gianini G, Granitzer M, Ziegler K (2018) Analysing neural network topologies: a game theoretic approach. Proc Comput Sci 126:234–243
Sughasiny M, Rajeshwari J (2018) Application of machine learning techniques, big data analytics in health care sector—a literature survey. In: 2018 2nd International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud)(I-SMAC) I-SMAC (IoT in Social, Mobile, Analytics and Cloud)(I-SMAC), 2018 2nd International Conference on, pp 741–749. IEEE
Szczepaniak PS, Lisboa PJ (2012) Fuzzy systems in medicine. Physica 41
Walia N, Singh H, Sharma A (2015) Anfis: adaptive neuro-fuzzy inference system-a survey. Int J Comput Appl 123(13)
Wang T, Yang L, Liu Q (2018) Beetle swarm optimization algorithm: theory and application. arXiv preprint arXiv:1808.00206
Zadeh LA (1996) Fuzzy sets. In: Fuzzy sets, fuzzy logic, and fuzzy systems: selected papers by Lotfi A Zadeh, pp 394–432. World Scientific
Acknowledgements
This work was partially supported by the European Commission through the project Smart Bear (Programme H2020-SC1-FA-DTS-2018-2, Grant Agreement 857172) and by the Lombardy Region (Italy) through the project MINDFoodsHub funded by the PON FESR 2014-2020 (Project ID 1176436).
Funding
Open access funding provided by Università degli Studi di Milano within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in the published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Singh, P., Kaur, A., Batth, R.S. et al. Multi-disease big data analysis using beetle swarm optimization and an adaptive neuro-fuzzy inference system. Neural Comput & Applic 33, 10403–10414 (2021). https://doi.org/10.1007/s00521-021-05798-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-021-05798-x