Abstract
In the fields of pattern recognition and machine learning, the use of data preprocessing algorithms has been increasing in recent years to achieve high classification performance. In particular, it has become inevitable to use the data preprocessing method prior to classification algorithms in classifying medical datasets with the nonlinear and imbalanced data distribution. In this study, a new data preprocessing method has been proposed for the classification of Parkinson, hepatitis, Pima Indians, single proton emission computed tomography (SPECT) heart, and thoracic surgery medical datasets with the nonlinear and imbalanced data distribution. These datasets were taken from UCI machine learning repository. The proposed data preprocessing method consists of three steps. In the first step, the cluster centers of each attribute were calculated using k-means, fuzzy c-means, and mean shift clustering algorithms in medical datasets including Parkinson, hepatitis, Pima Indians, SPECT heart, and thoracic surgery medical datasets. In the second step, the absolute differences between the data in each attribute and the cluster centers are calculated, and then, the average of these differences is calculated for each attribute. In the final step, the weighting coefficients are calculated by dividing the mean value of the difference to the cluster centers, and then, weighting is performed by multiplying the obtained weight coefficients by the attribute values in the dataset. Three different attribute weighting methods have been proposed: (1) similarity-based attribute weighting in k-means clustering, (2) similarity-based attribute weighting in fuzzy c-means clustering, and (3) similarity-based attribute weighting in mean shift clustering. In this paper, we aimed to aggregate the data in each class together with the proposed attribute weighting methods and to reduce the variance value within the class. Thus, by reducing the value of variance in each class, we have put together the data in each class and at the same time, we have further increased the discrimination between the classes. To compare with other methods in the literature, the random subsampling has been used to handle the imbalanced dataset classification. After attribute weighting process, four classification algorithms including linear discriminant analysis, k-nearest neighbor classifier, support vector machine, and random forest classifier have been used to classify imbalanced medical datasets. To evaluate the performance of the proposed models, the classification accuracy, precision, recall, area under the ROC curve, κ value, and F-measure have been used. In the training and testing of the classifier models, three different methods including the 50–50% train–test holdout, the 60–40% train–test holdout, and tenfold cross-validation have been used. The experimental results have shown that the proposed attribute weighting methods have obtained higher classification performance than random subsampling method in the handling of classifying of the imbalanced medical datasets.

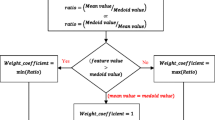
Reproduced with permission from [3]

















Similar content being viewed by others
References
Longadge R, Dongre SS, Malik L (2013) Class imbalance problem in data mining: review. Int J Comput Sci Netw (IJCSN) 2(1):83–87
https://classeval.wordpress.com. Accessed Feb 2018
http://sebastianraschka.com/Articles/2014_kernel_pca.html. Accessed Feb 2018
https://arxiv.org/ftp/arxiv/papers/1305/1305.1707. Accessed Feb 2018
Gong J, Kim H (2017) RHSBoost: improving classification performance in imbalance data. Comput Stat Data Anal 111:1–13
Shilaskar S, Ghato A, Chatur P (2017) Medical decision support system for extremely imbalanced datasets. Inf Sci 384:205–219
Zhang J, Xiao W, Li Y, Zhang S, Yang W (2017) Class-specific cost regulation extreme learning machine for imbalanced classification. Neurocomputing 261:70–82
UCI machine learning repository (2018) https://archive.ics.uci.edu/ml. Accessed Feb 2018
Short RD, Fukunaga K (1981) The optimal distance measure for nearest neighbor classification. IEEE Trans Inf Theory 27:622–627. https://doi.org/10.1109/TIT.1981.1056403
Weinberger KQ, Saul LK (2009) Distance metric learning for large margin nearest neighbor classification. J Mach Learn Res 10:207–244
Cost S, Salzberg S (1993) A weighted nearest neighbor algorithm for learning with symbolic attributes. Mach Learn 10:57–78. https://doi.org/10.1007/BF00993481
Breiman L (2001) Random forests. Mach Learn 45:5–32. https://doi.org/10.1023/A:1010933404324
Zhang Z (2014) Too much covariates in a multivariable model may cause the problem of overfitting. J Thorac Dis 6:E196–E197
Hastie T, Tibshirani R, Friedman JH (2003) The elements of statistical learning. Springer, New York
McLachlan GJ (2004) Discriminant analysis and statistical pattern recognition. Wiley, Hoboken
Shen R, Ghosh D, Chinnaiyan A, Meng Z (2006) Eigengene-based linear discriminant model for tumor classification using gene expression microarray data. Bioinformatics 22(21):2635–2642
Cortes C, Vapnik V (1995) Support-vector network. Mach Learn 20(3):273–297
Vapnik V (2014) Invited speaker. In: IPMU information processing and management. 15th international conference on information processing and management of uncertainty in knowledge-based systems, IPMU'2014, Montpellier, France,15–19 July 2014
Savitt JM, Dawson VL, Dawson TM (2006) Diagnosis and treatment of Parkinson disease: molecules to medicine. J Clin Investig 116(7):1744–1754. https://doi.org/10.1172/JCI29178
Levine CB, Fahrbach KR, Siderowf AD, Estok RP, Ludensky VM, Ross SD (2003) Diagnosis and treatment of Parkinson’s disease: a systematic review of the literature. Evid Rep Technol Assess 57:1–4 (Summary)
Yuvaraj R, Murugappan M, Acharya UR, Adeli H, Ibrahim NM, Mesquita E (2016) Brain functional connectivity patterns for emotional state classification in Parkinson’s disease patients without dementia. Behav Brain Res 298((Pt B)):248–260. https://doi.org/10.1016/j.bbr.2015.10.036
http://www.frank-dieterle.de/phd/2_4_3.html. Accessed Feb 2018
Clark M (2015) An introduction to machine learning with applications in R, Lecture notes. University of Notre Dame, Notre Dame
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that there is no conflict of interest in anywhere.
Rights and permissions
About this article
Cite this article
Polat, K. Similarity-based attribute weighting methods via clustering algorithms in the classification of imbalanced medical datasets. Neural Comput & Applic 30, 987–1013 (2018). https://doi.org/10.1007/s00521-018-3471-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-018-3471-8