Abstract
Background
Clostridium difficile infection (CDI) is the most common hospital-acquired infection. Unfortunately, genes that identify CDI-susceptible patients have not been well described. We performed a genome-wide association study (GWAS) to determine genetic variants associated with the development of CDI.
Methods
A cohort study of Caucasian patients undergoing autologous stem cell transplantation for multiple myeloma was performed. Patients were genotyped using Illumina® Whole Genome Genotyping Infinium chemistry. We then compared CDI-positive to CDI-negative patients using logistic regression for baseline clinical factors and false discovery rate (FDR) for genetic factors [single nucleotide polymorphisms (SNPs)]. SNPs associated with CDI at FDR of p < 0.01 were then incorporated into a logistic regression model combining clinical and genetic factors.
Results
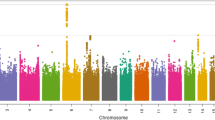
Of the 646 patients analyzed (59.7% male), 57 patients were tested CDI positive (cases) and were compared to 589 patients who were tested negative (controls). Hemoglobin, albumin, and hematocrit were lower for cases (p < 0.05). Eight SNPs on five genes (FLJ16171, GORASP2, RLBP1L1, ASPH, ATP7B) were associated with CDI at FDR p < 0.01. In the combined clinical and genetic model, low albumin and three genes RLBP1L1, ASPH, and ATP7B were associated with CDI.
Conclusion
Low serum albumin and genes RLBP1L1 and ASPH located on chromosome 8 and ATP7B on chromosome 13 were associated with CDI. Of particular interest is ATP7B given its copper modulatory role and the sporicidal properties of copper against Clostridium difficile.



Similar content being viewed by others
References
Scappaticci GB, Perissinotti AJ, Nagel JL, Bixby DL, Marini BL (2017) Risk factors and impact of clostridium difficile recurrence on haematology patients. J Antimicrob Chemother 72(5):1488–1495
Lessa FC, Gould CV, McDonald LC (2012) Current status of clostridium difficile infection epidemiology. Clin Infect Dis 55(Suppl 2):S65–S70
Bodet CA 3rd, Jorgensen JH, Drutz DJ (1985) Antibacterial activities of antineoplastic agents. Antimicrob Agents Chemother 28(3):437–439
Jain T, Croswell C, Urday-Cornejo V, Awali R, Cutright J, Salimnia H, Reddy Banavasi HV, Liubakka A, Lephart P, Chopra T, Revankar SG, Chandrasekar P, Alangaden G (2016) Clostridium difficile colonization in hematopoietic stem cell transplant recipients: a prospective study of the epidemiology and outcomes involving toxigenic and nontoxigenic strains. Biol Blood Marrow Transplant 22(1):157–163
Ananthakrishnan AN, Oxford EC, Nguyen DD, Sauk J, Yajnik V, Xavier RJ (2013) Genetic risk factors for clostridium difficile infection in ulcerative colitis. Aliment Pharmacol Ther 38(5):522–530
Mosteller RD (1987) Simplified calculation of body-surface area. N Engl J Med 317(17):1098
Krishna SG, Zhao W, Apewokin SK, Krishna K, Chepyala P, Anaissie EJ (2013) Risk factors, preemptive therapy, and antiperistaltic agents for clostridium difficile infection in cancer patients. Transpl Infect Dis 15(5):493–501
Armitage P (1955) Tests for linear trends in proportions and frequencies. Int Biom Soc 11(3):375–386
Benjamimi Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57(1):289–300
Apewokin S, Coleman E, Enderlin C, Goodwin J, Lee J, Erickson S, Mckelvey K, Raj V, Kumar NS, Daohong Z (2014) Genetic variants associated with the development of clostridium difficile infection during autologous stem cell transplantation. Open Forum Infect Dis 1(suppl 1):S166–S167
Brandt LJ (2015) Fecal microbiota transplant: respice, adspice, prospice. J Clin Gastroenterol 49(Suppl 1):S65–S68
Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, de Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB (2007) A whole-genome association study of major determinants for host control of HIV-1. Science 317(5840):944–947
Rauch A, Kutalik Z, Descombes P et al (2010) Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138(4):1338–1345 1345.e1–7
Seyfizadeh N, Seyfizadeh N, Gharibi T, Babaloo Z (2015) Interleukin-13 as an important cytokine: a review on its roles in some human diseases. Acta Microbiol Immunol Hung 62(4):341–378
Engevik MA, Yacyshyn MB, Engevik KA, Wang J, Darien B, Hassett DJ, Yacyshyn BR, Worrell RT (2015) Human clostridium difficile infection: altered mucus production and composition. Am J Physiol Gastrointest Liver Physiol 308(6):G510–G524
Fallon PG, Richardson EJ, McKenzie GJ, McKenzie AN (2000) Schistosome infection of transgenic mice defines distinct and contrasting pathogenic roles for IL-4 and IL-13: IL-13 is a profibrotic agent. J Immunol 164(5):2585–2591
Daugherty RL, Gottardi CJ (2007) Phospho-regulation of beta-catenin adhesion and signaling functions. Physiology (Bethesda) 22:303–309
Garey KW, Jiang ZD, Ghantoji S, Tam VH, Arora V, Dupont HL (2010) A common polymorphism in the interleukin-8 gene promoter is associated with an increased risk for recurrent clostridium difficile infection. Clin Infect Dis 51(12):1406–1410
Bull PC, Thomas GR, Rommens JM, Forbes JR, Cox DW (1993) The Wilson disease gene is a putative copper transporting P-type ATPase similar to the menkes gene. Nat Genet 5(4):327–337
Park HD, Park HK, Chung HS, Lee SY, Kim JW, Ki CS (2010) Association of ATP7B mutation detection rate with biochemical characteristics in korean patients with Wilson disease. Ann Clin Lab Sci 40(1):15–19
Weaver L, Michels HT, Keevil CW (2008) Survival of clostridium difficile on copper and steel: futuristic options for hospital hygiene. J Hosp Infect 68(2):145–151
O'Gorman J, Humphreys H (2012) Application of copper to prevent and control infection. Where are we now? J Hosp Infect 81(4):217–223
Sorg JA (2016) Microbial bile acid metabolic clusters: the bouncers at the bar. Cell Host Microbe 16:551–552
Katoh Y, Ritter B, Gaffry T, Blondeau F, Honing S, McPherson PS (2009) The clavesin family, neuron-specific lipid- and clathrin-binding Sec14 proteins regulating lysosomal morphology. J Biol Chem 284(40):27646–27654
Polishchuk EV, Concilli M, Iacobacci S, Chesi G, Pastore N, Piccolo P, Paladino S, Baldantoni D, van IJzendoorn SCD, Chan J, Chang CJ, Amoresano A, Pane F, Pucci P, Tarallo A, Parenti G, Brunetti-Pierri N, Settembre C, Ballabio A, Polishchuk RS (2014) Wilson disease protein ATP7B utilizes lysosomal exocytosis to maintain copper homeostasis. Dev Cell 29(6):686–700
Korioth F, Gieffers C, Frey J (1994) Cloning and characterization of the human gene encoding aspartyl beta-hydroxylase. Gene 150(2):395–399
Kochan TJ, Somers MJ, Kaiser AM, Shoshiev MS, Hagan AK, Hastie JL, Giordano NP, Smith AD, Schubert AM, Carlson PE, Hanna PC (2017) Intestinal calcium and bile salts facilitate germination of clostridium difficile spores. PLoS Pathog 13(7):e1006443
Acknowledgements
Many thanks to Xenofon Papanicolau and Tesfaye Mersha for help with Pathway analysis.
Funding
Financial support to Elizabeth Ann Coleman was provided by the National Institutes of Health (NIH)/National Institute of Nursing Research (NINR) 5 RC2NR011945, and for additional support, the Translational Research Institute at UAMS (grant no. 1UL1RR029884) and the Elizabeth Stanley Cooper Chair in Oncology Nursing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
ESM 1
(XLSX 32629 kb)
Rights and permissions
About this article
Cite this article
Apewokin, S., Lee, J.Y., Goodwin, J.A. et al. Host genetic susceptibility to Clostridium difficile infections in patients undergoing autologous stem cell transplantation: a genome-wide association study. Support Care Cancer 26, 3127–3134 (2018). https://doi.org/10.1007/s00520-018-4173-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00520-018-4173-6