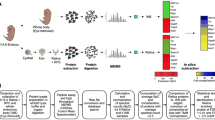
Abstract
Isolated or syndromic congenital cataracts are heterogeneous developmental defects, making the identification of the associated genes challenging. In the past, mouse lens expression microarrays have been successfully applied in bioinformatics tools (e.g., iSyTE) to facilitate human cataract-associated gene discovery. To develop a new resource for geneticists, we report high-throughput RNA sequencing (RNA-seq) profiles of mouse lens at key embryonic stages (E)10.5 (lens pit), E12.5 (primary fiber cell differentiation), E14.5 and E16.5 (secondary fiber cell differentiation). These stages capture important events as the lens develops from an invaginating placode into a transparent tissue. Previously, in silico whole-embryo body (WB)-subtraction-based “lens-enriched” expression has been effective in prioritizing cataract-linked genes. To apply an analogous approach, we generated new mouse WB RNA-seq datasets and show that in silico WB subtraction of lens RNA-seq datasets successfully identifies key genes based on lens-enriched expression. At ≥2 counts-per-million expression, ≥1.5 log2 fold-enrichment (p < 0.05) cutoff, E10.5 lens exhibits 1401 enriched genes (17% lens-expressed genes), E12.5 lens exhibits 1937 enriched genes (22% lens-expressed genes), E14.5 lens exhibits 2514 enriched genes (31% lens-expressed genes), and E16.5 lens exhibits 2745 enriched genes (34% lens-expressed genes). Biological pathway analysis identified genes associated with lens development, transcription regulation and signaling pathways, among other functional groups. Furthermore, these new RNA-seq data confirmed high expression of established cataract-linked genes and identified new potential regulators in the lens. Finally, we developed new lens stage-specific UCSC Genome Brower annotation tracks and made these publicly accessible through iSyTE (https://research.bioinformatics.udel.edu/iSyTE/) for user-friendly visualization of lens gene expression/enrichment to prioritize genes from high-throughput data from cataract cases.







Similar content being viewed by others
References
Agrawal SA, Anand D, Siddam AD et al (2015) Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum Genet 134:717–735. https://doi.org/10.1007/s00439-015-1554-5
Aldahmesh MA, Khan AO, Mohamed JY et al (2012) Genomic analysis of pediatric cataract in Saudi Arabia reveals novel candidate disease genes. Genet Med 14:955–962. https://doi.org/10.1038/gim.2012.86
Aldahmesh MA, Alshammari MJ, Khan AO et al (2013) The syndrome of microcornea, myopic chorioretinal atrophy, and telecanthus (MMCAT) is caused by mutations in ADAMTS18. Hum Mutat 34:1195–1199. https://doi.org/10.1002/humu.22374
Anand D, Lachke SA (2017) Systems biology of lens development: a paradigm for disease gene discovery in the eye. Exp Eye Res 156:22–33. https://doi.org/10.1016/j.exer.2016.03.010
Anand D, Agrawal S, Siddam A et al (2015) An integrative approach to analyze microarray datasets for prioritization of genes relevant to lens biology and disease. Genomics Data 5:223–227. https://doi.org/10.1016/j.gdata.2015.06.017
Ashery-Padan R, Marquardt T, Zhou X, Gruss P (2000) Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev 14:2701–2711
Audette DS, Anand D, So T et al (2016) Prox1 and fibroblast growth factor receptors form a novel regulatory loop controlling lens fiber differentiation and gene expression. Development 143:318–328. https://doi.org/10.1242/dev.127860
Bassnett S, Shi Y, Vrensen GFJM (2011) Biological glass: structural determinants of eye lens transparency. Philos Trans R Soc Lond B Biol Sci 366:1250–1264. https://doi.org/10.1098/rstb.2010.0302
Blackshaw S, Fraioli RE, Furukawa T, Cepko CL (2001) Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell 107:579–589
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Brennan LA, Kantorow WL, Chauss D et al (2012) Spatial expression patterns of autophagy genes in the eye lens and induction of autophagy in lens cells. Mol Vis 18:1773–1786
Budak G, Dash S, Srivastava R et al (2018) Express: a database of transcriptome profiles encompassing known and novel transcripts across multiple development stages in eye tissues. Exp Eye Res 168:57–68. https://doi.org/10.1016/j.exer.2018.01.009
Cavalheiro GR, Matos-Rodrigues GE, Zhao Y et al (2017) N-myc regulates growth and fiber cell differentiation in lens development. Dev Biol 429:105–117. https://doi.org/10.1016/j.ydbio.2017.07.002
Chauhan BK, Reed NA, Yang Y et al (2002a) A comparative cDNA microarray analysis reveals a spectrum of genes regulated by Pax6 in mouse lens. Genes Cells 7:1267–1283
Chauhan BK, Reed NA, Zhang W et al (2002b) Identification of genes downstream of Pax6 in the mouse lens using cDNA microarrays. J Biol Chem 277:11539–11548. https://doi.org/10.1074/jbc.M110531200
Chograni M, Alkuraya FS, Ourteni I et al (2015) Autosomal recessive congenital cataract, intellectual disability phenotype linked to STX3 in a consanguineous Tunisian family. Clin Genet 88:283–287. https://doi.org/10.1111/cge.12489
Cvekl A, Zhang X (2017) Signaling and gene regulatory networks in mammalian lens development. Trends Genet 33:677–702. https://doi.org/10.1016/j.tig.2017.08.001
Dash S, Dang CA, Beebe DC, Lachke SA (2015) Deficiency of the RNA binding protein Caprin2 causes lens defects and features of Peters anomaly. Dev Dyn. https://doi.org/10.1002/dvdy.24303
David D, Anand D, Araújo C et al (2018) Identification of OAF and PVRL1 as candidate genes for an ocular anomaly characterized by Peters anomaly type 2 and ectopia lentis. Exp Eye Res 168:161–170. https://doi.org/10.1016/j.exer.2017.12.012
De Maria A, Bassnett S (2015) Birc7: a late fiber gene of the crystalline lens. Investig Ophthalmol Vis Sci 56:4823–4834. https://doi.org/10.1167/iovs.15-16968
Evers C, Paramasivam N, Hinderhofer K et al (2015) SIPA1L3 identified by linkage analysis and whole-exome sequencing as a novel gene for autosomal recessive congenital cataract. Eur J Hum Genet 23:1627–1633. https://doi.org/10.1038/ejhg.2015.46
Greenlees R, Mihelec M, Yousoof S et al (2015) Mutations in SIPA1L3 cause eye defects through disruption of cell polarity and cytoskeleton organization. Hum Mol Genet 24:5789–5804. https://doi.org/10.1093/hmg/ddv298
Greiling TMS, Stone B, Clark JI (2009) Absence of SPARC leads to impaired lens circulation. Exp Eye Res 89:416–425. https://doi.org/10.1016/j.exer.2009.04.008
Hawse JR, Hejtmancik JF, Huang Q et al (2003) Identification and functional clustering of global gene expression differences between human age-related cataract and clear lenses. Mol Vis 9:515–537
Hawse JR, Hejtmancik JF, Horwitz J, Kantorow M (2004) Identification and functional clustering of global gene expression differences between age-related cataract and clear human lenses and aged human lenses. Exp Eye Res 79:935–940. https://doi.org/10.1016/j.exer.2004.04.007
Hawse JR, DeAmicis-Tress C, Cowell TL, Kantorow M (2005) Identification of global gene expression differences between human lens epithelial and cortical fiber cells reveals specific genes and their associated pathways important for specialized lens cell functions. Mol Vis 11:274–283
Hoang TV, Kumar PKR, Sutharzan S et al (2014) Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing. Mol Vis 20:1491–1517
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. https://doi.org/10.1038/nprot.2008.211
Ivanov D, Dvoriantchikova G, Pestova A et al (2005) Microarray analysis of fiber cell maturation in the lens. FEBS Lett 579:1213–1219. https://doi.org/10.1016/j.febslet.2005.01.016
Kakrana A, Yang A, Anand D et al (2018) iSyTE 2.0: a database for expression-based gene discovery in the eye. Nucleic Acids Res 46:D875–D885. https://doi.org/10.1093/nar/gkx837
Kasaikina MV, Fomenko DE, Labunskyy VM et al (2011) Roles of the 15-kDa selenoprotein (Sep15) in redox homeostasis and cataract development revealed by the analysis of Sep 15 knockout mice. J Biol Chem 286:33203–33212. https://doi.org/10.1074/jbc.M111.259218
Khan SY, Hackett SF, Lee M-CW et al (2015) Transcriptome profiling of developing murine lens through RNA sequencing. Investig Ophthalmol Vis Sci 56:4919–4926. https://doi.org/10.1167/iovs.14-16253
Khan SY, Hackett SF, Riazuddin SA (2016) Non-coding RNA profiling of the developing murine lens. Exp Eye Res 145:347–351. https://doi.org/10.1016/j.exer.2016.01.010
Khan SY, Ali M, Kabir F et al (2018) Identification of novel transcripts and peptides in developing murine lens. Sci Rep. https://doi.org/10.1038/s41598-018-28727-w
Krall M, Htun S, Anand D et al (2018) A zebrafish model of foxe3 deficiency demonstrates lens and eye defects with dysregulation of key genes involved in cataract formation in humans. Hum Genet 137:315–328. https://doi.org/10.1007/s00439-018-1884-1
Lachke SA, Alkuraya FS, Kneeland SC et al (2011) Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science 331:1571–1576. https://doi.org/10.1126/science.1195970
Lachke SA, Higgins AW, Inagaki M et al (2012a) The cell adhesion gene PVRL3 is associated with congenital ocular defects. Hum Genet 131:235–250. https://doi.org/10.1007/s00439-011-1064-z
Lachke SA, Ho JWK, Kryukov GV et al (2012b) iSyTE: integrated systems tool for eye gene discovery. Investig Ophthalmol Vis Sci 53:1617–1627. https://doi.org/10.1167/iovs.11-8839
Liu W, Lagutin OV, Mende M et al (2006) Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J 25:5383–5395. https://doi.org/10.1038/sj.emboj.7601398
Manthey AL, Lachke SA, FitzGerald PG et al (2014a) Loss of Sip1 leads to migration defects and retention of ectodermal markers during lens development. Mech Dev 131:86–110. https://doi.org/10.1016/j.mod.2013.09.005
Manthey AL, Terrell AM, Lachke SA et al (2014b) Development of novel filtering criteria to analyze RNA-sequencing data obtained from the murine ocular lens during embryogenesis. Genomics Data 2:369–374. https://doi.org/10.1016/j.gdata.2014.10.015
Patel N, Khan AO, Mansour A et al (2014) Mutations in ASPH cause facial dysmorphism, lens dislocation, anterior-segment abnormalities, and spontaneous filtering blebs, or Traboulsi syndrome. Am J Hum Genet 94:755–759. https://doi.org/10.1016/j.ajhg.2014.04.002
Patel N, Anand D, Monies D et al (2017) Novel phenotypes and loci identified through clinical genomics approaches to pediatric cataract. Hum Genet 136:205–225. https://doi.org/10.1007/s00439-016-1747-6
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Rothe M, Kanwal N, Dietmann P et al (2017) An Epha4/Sipa1l3/Wnt pathway regulates eye development and lens maturation. Development 144:321–333. https://doi.org/10.1242/dev.147462
Rowan S, Conley KW, Le TT et al (2008) Notch signaling regulates growth and differentiation in the mammalian lens. Dev Biol 321:111–122. https://doi.org/10.1016/j.ydbio.2008.06.002
Saravanamuthu SS, Le TT, Gao CY et al (2012) Conditional ablation of the Notch2 receptor in the ocular lens. Dev Biol 362:219–229. https://doi.org/10.1016/j.ydbio.2011.11.011
Siddam AD, Gautier-Courteille C, Perez-Campos L et al (2018) The RNA-binding protein Celf1 post-transcriptionally regulates p27Kip1 and Dnase2b to control fiber cell nuclear degradation in lens development. PLoS Genet 14:e1007278. https://doi.org/10.1371/journal.pgen.1007278
Sousounis K, Tsonis PA (2012) Patterns of gene expression in microarrays and expressed sequence tags from normal and cataractous lenses. Hum Genomics 6:14. https://doi.org/10.1186/1479-7364-6-14
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-SEq. Bioinformatics 25:1105–1111. https://doi.org/10.1093/bioinformatics/btp120
Trapnell C, Roberts A, Goff L et al (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. https://doi.org/10.1038/nprot.2012.016
Wang Y, Terrell AM, Riggio BA et al (2017) β1-integrin deletion from the lens activates cellular stress responses leading to apoptosis and fibrosis. Investig Ophthalmol Vis Sci 58:3896–3922. https://doi.org/10.1167/iovs.17-21721
Wolf L, Gao CS, Gueta K et al (2013a) Identification and characterization of FGF2-dependent mRNA: microRNA networks during lens fiber cell differentiation. G3 (Bethesda) 3:2239–2255. https://doi.org/10.1534/g3.113.008698
Wolf L, Harrison W, Huang J et al (2013b) Histone posttranslational modifications and cell fate determination: lens induction requires the lysine acetyltransferases CBP and p300. Nucleic Acids Res 41:10199–10214. https://doi.org/10.1093/nar/gkt824
Xiao W, Liu W, Li Z et al (2006) Gene expression profiling in embryonic mouse lenses. Mol Vis 12:1692–1698
Zhang H-M, Liu T, Liu C-J et al (2015) AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res 43:D76–D81. https://doi.org/10.1093/nar/gku887
Zhang Y, Fan J, Ho JWK et al (2016) Crim1 regulates integrin signaling in murine lens development. Development 143:356–366. https://doi.org/10.1242/dev.125591
Zhao Y, Zheng D, Cvekl A (2018) A comprehensive spatial-temporal transcriptomic analysis of differentiating nascent mouse lens epithelial and fiber cells. Exp Eye Res 175:56–72. https://doi.org/10.1016/j.exer.2018.06.004
Acknowledgements
This work was supported by the National Institutes of Health awards R01EY021505 (S.L.) from National Eye Institute, and R03DE024776 (S.L., I.S.) from National Institute of Dental and Craniofacial Research. We thank the University of Kansas Medical Center Genomics Core for generating the sequence data sets. The Genomics Core is supported by the University of Kansas School of Medicine, the Kansas Intellectual and Developmental Disability Research Center (NIH U54 HD090216) and the Molecular Regulation of Cell Development and Differentiation COBRE (5P20GM104936).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Anand, D., Kakrana, A., Siddam, A.D. et al. RNA sequencing-based transcriptomic profiles of embryonic lens development for cataract gene discovery. Hum Genet 137, 941–954 (2018). https://doi.org/10.1007/s00439-018-1958-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-018-1958-0