Abstract
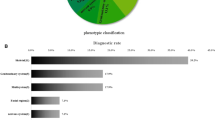
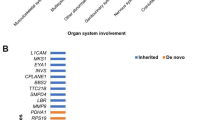
Whole exome sequencing (WES) is an emerging technique in prenatal diagnosis. In this retrospective study, we examined diagnostic utility and limitations of WES in prenatal cases with structural birth defects. DNA from 20 trios (fetal and parental), with normal karyotype and microarray findings, underwent WES and variant interpretation at a reference laboratory. The WES results were later re-evaluated in our academic center utilizing prenatal and postnatal phenotyping. Initial analysis using only prenatal ultrasound findings revealed no pathogenic or likely pathogenic variants in 20 pregnancies with structural birth defects. Re-analysis of WES variants and combination of prenatal and postnatal phenotyping yielded pathogenic variants in at least 20% of cases including PORCN gene in a fetus with split-hand/foot malformation, as well as variants of uncertain significance in NEB and NOTCH1 in fetuses with postnatal muscle weakness and Adams–Oliver syndrome, respectively. Furthermore, Sanger sequencing in a patient with holoprosencephaly, elucidated by postnatal MRI, revealed a pathogenic 47-base pairs deletion in ZIC2 which was missed by prenatal WES. This study suggests that incomplete prenatal phenotyping and lack of prenatal ultrasound-genotype databases are the limiting factors for current interpretation of WES data in prenatal diagnosis. Development of prenatal phenotype–genotype databases would significantly help WES interpretation in this setting. Patients who underwent prenatal clinical WES may benefit from the re-analysis based on detailed postnatal findings.
Similar content being viewed by others
References
Abou Tayoun AN, Spinner NB, Rehm HL, Green RC, Bianchi DW (2017) Prenatal DNA sequencing: clinical, counseling, and diagnostic laboratory considerations. Prenat Diagn. https://doi.org/10.1002/pd.5038
Alamillo CL, Powis Z, Farwell K, Shahmirzadi L, Weltmer EC, Turocy J, Lowe T, Kobelka C, Chen E, Basel D, Ashkinadze E, D’Augelli L, Chao E, Tang S (2015) Exome sequencing positively identified relevant alterations in more than half of cases with an indication of prenatal ultrasound anomalies. Prenat Diagn 35(11):1073–1078. https://doi.org/10.1002/pd.4648
American College of Obstetricians and Gynecologists (ACOG), Committee on Genetics, the Society for Maternal-Fetal Medicine (2016) ACOG Committee Opinion No. 682: microarrays and next-generation sequencing technology: the use of advanced genetic diagnostic tools in obstetrics and gynecology. Obstet Gynecol 128(6):e262–e268. https://doi.org/10.1097/aog.0000000000001817
Brady PD, Delle Chiaie B, Christenhusz G, Dierickx K, Van Den Bogaert K, Menten B, Janssens S, Defoort P, Roets E, Sleurs E, Keymolen K, De Catte L, Deprest J, de Ravel T, Van Esch H, Fryns JP, Devriendt K, Vermeesch JR (2014) A prospective study of the clinical utility of prenatal chromosomal microarray analysis in fetuses with ultrasound abnormalities and an exploration of a framework for reporting unclassified variants and risk factors. Genet Med 16(6):469–476. https://doi.org/10.1038/gim.2013.168
Brady PD, Van Esch H, Fieremans N, Froyen G, Slavotinek A, Deprest J, Devriendt K, Vermeesch JR (2015) Expanding the phenotypic spectrum of PORCN variants in two males with syndromic microphthalmia. Eur J Hum Genet 23(4):551–554. https://doi.org/10.1038/ejhg.2014.135
Bulas D, Egloff A (2013) Benefits and risks of MRI in pregnancy. Semin Perinatol 37(5):301–304. https://doi.org/10.1053/j.semperi.2013.06.005
Carss KJ, Hillman SC, Parthiban V, McMullan DJ, Maher ER, Kilby MD, Hurles ME (2014) Exome sequencing improves genetic diagnosis of structural fetal abnormalities revealed by ultrasound. Hum Mol Genet 23(12):3269–3277. https://doi.org/10.1093/hmg/ddu038
Centers for Disease Control and Prevention (2008) Update on overall prevalence of major birth defects—Atlanta, Georgia, 1978–2005. Morb Mortal Wkly Rep 57(1):1–5
Drury S, Williams H, Trump N, Boustred C, Gosgene Lench N, Scott RH, Chitty LS (2015) Exome sequencing for prenatal diagnosis of fetuses with sonographic abnormalities. Prenat Diagn 35(10):1010–1017. https://doi.org/10.1002/pd.4675
Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O’Daniel JM, Ormond KE, Rehm HL, Watson MS, Williams MS, Biesecker LG, American College of Medical Genetics and Genomics (2013) ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 15(7):565–574. https://doi.org/10.1038/gim.2013.73
Haque IS, Lazarin GA, Kang HP, Evans EA, Goldberg JD, Wapner RJ (2016) Modeled fetal risk of genetic diseases identified by expanded carrier screening. JAMA 316(7):734–742. https://doi.org/10.1001/jama.2016.11139
Kalynchuk EJ, Althouse A, Parker LS, Saller DN Jr, Rajkovic A (2015) Prenatal whole-exome sequencing: parental attitudes. Prenat Diagn 35(10):1030–1036. https://doi.org/10.1002/pd.4635
Kerstjens-Frederikse WS, van de Laar IM, Vos YJ, Verhagen JM, Berger RM, Lichtenbelt KD, Klein Wassink-Ruiter JS, van der Zwaag PA, du Marchie Sarvaas GJ, Bergman KA, Bilardo CM, Roos-Hesselink JW, Janssen JH, Frohn-Mulder IM, van Spaendonck-Zwarts KY, van Melle JP, Hofstra RM, Wessels MW (2016) Cardiovascular malformations caused by NOTCH1 mutations do not keep left: data on 428 probands with left-sided CHD and their families. Genet Med 18(9):914–923. https://doi.org/10.1038/gim.2015.193
Koeberl DD, McGillivray B, Sybert VP (1995) Prenatal diagnosis of 45,X/46,XX mosaicism and 45,X: implications for postnatal outcome. Am J Hum Genet 57(3):661–666
Mary L, Scheidecker S, Kohler M, Lombardi MP, Delezoide AL, Auberger E, Triau S, Colin E, Gerard M, Grzeschik KH, Dollfus H, Antal MC (2017) Prenatal diagnosis of focal dermal hypoplasia: report of three fetuses and review of the literature. Am J Med Genet A 173(2):479–486. https://doi.org/10.1002/ajmg.a.37974
Reddy UM, Page GP, Saade GR, Silver RM, Thorsten VR, Parker CB, Pinar H, Willinger M, Stoll BJ, Heim-Hall J, Varner MW, Goldenberg RL, Bukowski R, Wapner RJ, Drews-Botsch CD, O’Brien BM, Dudley DJ, Levy B, Network NSCR (2012) Karyotype versus microarray testing for genetic abnormalities after stillbirth. N Engl J Med 367(23):2185–2193. https://doi.org/10.1056/NEJMoa1201569
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Shamseldin HE, Kurdi W, Almusafri F, Alnemer M, Alkaff A, Babay Z, Alhashem A, Tulbah M, Alsahan N, Khan R, Sallout B, Al Mardawi E, Seidahmed MZ, Meriki N, Alsaber Y, Qari A, Khalifa O, Eyaid W, Rahbeeni Z, Kurdi A, Hashem M, Alshidi T, Al-Obeid E, Abdulwahab F, Ibrahim N, Ewida N, El-Akouri K, Al Mulla M, Ben-Omran T, Pergande M, Cirak S, Al Tala S, Shaheen R, Faqeih E, Alkuraya FS (2017) Molecular autopsy in maternal-fetal medicine. Genet Med. https://doi.org/10.1038/gim.2017.111
Singleton MV, Guthery SL, Voelkerding KV, Chen K, Kennedy B, Margraf RL, Durtschi J, Eilbeck K, Reese MG, Jorde LB, Huff CD, Yandell M (2014) Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am J Hum Genet 94(4):599–610. https://doi.org/10.1016/j.ajhg.2014.03.010
Solomon BD, Lacbawan F, Mercier S, Clegg NJ, Delgado MR, Rosenbaum K, Dubourg C, David V, Olney AH, Wehner LE, Hehr U, Bale S, Paulussen A, Smeets HJ, Hardisty E, Tylki-Szymanska A, Pronicka E, Clemens M, McPherson E, Hennekam RC, Hahn J, Stashinko E, Levey E, Wieczorek D, Roeder E, Schell-Apacik CC, Booth CW, Thomas RL, Kenwrick S, Cummings DA, Bous SM, Keaton A, Balog JZ, Hadley D, Zhou N, Long R, Velez JI, Pineda-Alvarez DE, Odent S, Roessler E, Muenke M (2010) Mutations in ZIC2 in human holoprosencephaly: description of a novel ZIC2 specific phenotype and comprehensive analysis of 157 individuals. J Med Genet 47(8):513–524. https://doi.org/10.1136/jmg.2009.073049
Stittrich AB, Lehman A, Bodian DL, Ashworth J, Zong Z, Li H, Lam P, Khromykh A, Iyer RK, Vockley JG, Baveja R, Silva ES, Dixon J, Leon EL, Solomon BD, Glusman G, Niederhuber JE, Roach JC, Patel MS (2014) Mutations in NOTCH1 cause Adams–Oliver syndrome. Am J Hum Genet 95(3):275–284. https://doi.org/10.1016/j.ajhg.2014.07.011
Vora NL, Powell B, Brandt A, Strande N, Hardisty E, Gilmore K, Foreman AKM, Wilhelmsen K, Bizon C, Reilly J, Owen P, Powell CM, Skinner D, Rini C, Lyerly AD, Boggess KA, Weck K, Berg JS, Evans JP (2017) Prenatal exome sequencing in anomalous fetuses: new opportunities and challenges. Genet Med 19(11):1207–1216. https://doi.org/10.1038/gim.2017.33
Wang X, Reid Sutton V, Omar Peraza-Llanes J, Yu Z, Rosetta R, Kou YC, Eble TN, Patel A, Thaller C, Fang P, Van den Veyver IB (2007) Mutations in X-linked PORCN, a putative regulator of Wnt signaling, cause focal dermal hypoplasia. Nat Genet 39(7):836–838. https://doi.org/10.1038/ng2057
Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, Hardison M, Person R, Bekheirnia MR, Leduc MS, Kirby A, Pham P, Scull J, Wang M, Ding Y, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, Eng CM (2013) Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 369(16):1502–1511. https://doi.org/10.1056/NEJMoa1306555
Yang L, Chen J, Shen B (2017) Newborn screening in the era of precision medicine. Adv Exp Med Biol 1005:47–61. https://doi.org/10.1007/978-981-10-5717-5_3
Yates CL, Monaghan KG, Copenheaver D, Retterer K, Scuffins J, Kucera CR, Friedman B, Richard G, Juusola J (2017) Whole-exome sequencing on deceased fetuses with ultrasound anomalies: expanding our knowledge of genetic disease during fetal development. Genet Med 19(10):1171–1178. https://doi.org/10.1038/gim.2017.31
Yatsenko SA, Davis S, Hendrix NW, Surti U, Emery S, Canavan T, Speer P, Hill L, Clemens M, Rajkovic A (2013) Application of chromosomal microarray in the evaluation of abnormal prenatal findings. Clin Genet 84(1):47–54. https://doi.org/10.1111/cge.12027
Acknowledgements
We thank the families for participation in this study.
Funding
This study was supported (to AR) by Magee-Womens Research Institute and Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This study was approved by the University of Pittsburgh Institutional Review Board (#PRO16070068).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Data availability
All the genetic and genomic data generated during this study are included in this manuscript and its supplementary files.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Aarabi, M., Sniezek, O., Jiang, H. et al. Importance of complete phenotyping in prenatal whole exome sequencing. Hum Genet 137, 175–181 (2018). https://doi.org/10.1007/s00439-017-1860-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-017-1860-1