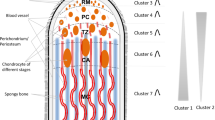
Abstract
Velvet antler displays the fastest and most robust tissue proliferation in the animal world, it is a model for a complete organ development/regeneration, and alternative medicine, tonic made from velvet antler, was beneficial for human. The weight of velvet antler had high biomedical and economic value, but the related regulation mechanisms controlling velvet antler weight remain unclear. In this study, extremely heavy and light velvet antler groups were selected from a sika deer population of 100 individuals with extreme velvet antler weight. A combination of full-length transcriptome sequencing and microRNA sequencing to the proliferation zone in the tip of velvet antler was applied. A total of 55306 transcripts and 1082 microRNAs were identified. Some highly expressed genes (COL1A1, COL1A2, COL3A1, FN1, and ATP6) and microRNAs (miR-21, let-7i, and miR-27b) were highly correlated with the physiological and growth characteristics of velvet antlers. Among the 334 differentially expressed genes, we found that most of the genes were located in the developmental process, especially animal organ development process. It is exciting to see that more blood vessels were found in the growing tip of heavy velvet antler through histological observation, and GO term of blood vessel development was also significant different between two groups. The combination analysis with mRNA and microRNA data in velvet antler showed a specific regulation network involved in the development of bone, mesenchyme, cartilage, and blood vessel, and helped us clearly find out the candidate 14 genes and 6 microRNAs, which could be used for selecting significant DNA markers of velvet antler weight.







Similar content being viewed by others
Data availability
All raw sequence reads were deposited in NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra) with SRA accession: SRP145255.
References
Berg DK, Li C, Asher G, Wells DN, Oback B (2007) Red deer cloned from antler stem cells and their differentiated progeny. Biol Reprod 77:384–394
Borsy A, Podani J, Steger V, Balla B, Horvath A, Kosa JP, Gyurjan I, Molnar A, Szabolcsi Z, Szabo L, Jako E, Zomborszky Z, Nagy J, Semsey S, Vellai T, Lakatos P, Orosz L (2009) Identifying novel genes involved in both deer physiological and human pathological osteoporosis. Mol Genet Genomics 281:301–313
Clark DE, Li CY, Wang WY, Martin SK, Suttie JM (2006) Vascular localization and proliferation in the growing tip of the deer antler. Anat Rec Part A 288A:973–981
Dale R. McCullough ST, Koichi K (2009) Sika Deer: biology and management of native and introduced populations [M]. Springer, Japan
Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ (2008) The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 7:759–764
Francis SM, Suttie JM (1998) Detection of growth factors and proto-oncogene mRNA in the growing tip of red deer (Cervus elaphus) antler using reverse-transcriptase polymerase chain reaction (RT-PCR). J Exp Zool 281:36–42
Gao LA, Tao DY, Shan YC, Liang Z, Zhang LH, Huo YS, Zhang YK (2010) HPLC-MS/MS shotgun proteomic research of deer antlers with multiparallel protein extraction methods. J Chromatogr B: Anal Technol Biomed Life Sci 878:3370–3374
Goss RJ (1983) Deer antlers: regeneration, function and evolution [M]. Academic Press, New York
Hao L, Li HP, Yan L (2011) [Analysis of expressed sequence tags (ESTs) in sika deer (Cervus nippon hortulorum) velvet tip tissue]. Yi Chuan 33:371–377
Hu PF, Xu JP, Ai C, Shao XJ, Wang HL, Dong YM, Cui XZ, Fuhe Y, Xiumei X (2017) Screening weight related genes of velvet antlers by whole genome re-sequencing. Yi Chuan 39:1090–1101
Huang FJ, Ji JX, Qian J, Yang KY, Zhang Y, Xin YQ, Jing ZG, Wang MH, Chen TY, Wu WT (2010) Isolation, characterization and biological activity of a novel peptide from sika (Cervus nippon Temminck) antler. Pharm Biotechnol 17:151–156
Jia BY, Ba HX, Wang GW, Yang Y, Cui XZ, Peng YH, Zheng JJ, Xing XM, Yang FH (2016) Transcriptome analysis of sika deer in China. Mol Genet Genomics 291:1941–1953
Kruuk LEB, Slate J, Pemberton JM, Brotherstone S, Guinness F, Clutton-Brock T (2002) Antler size in red deer: Heritability and selection but no evolution. Evolution 56:1683–1695
Li H (2003) Study on the performance of the velvet antler of Sika Deer. Zhongguo Xumu Zazhi 39:31–32
Li HPZX, Bing GL, Li S, Zhao M, Chen XJ (1997) Specific character analysis on artificial breed and strain of deer in China. Yichuan 19:76–78
Li CY, Yang FH, Sheppard A (2009) Adult stem cells and mammalian epimorphic regeneration-insights from studying annual renewal of deer antlers. Curr Stem Cell Res Ther 4:237–251
Li C, Harper A, Puddick J, Wang W, McMahon C (2012) Proteomes and signalling pathways of antler stem cells. PLoS One 7:e30026. https://doi.org/10.1371/journal.pone.0030026
Liu M (2013) Expression changes of classical signal transduction system during velvet antler growth and development[D]. Changchun Univ Chin Med
Mount JG, Muzylak M, Allen S, Althnaian T, McGonnell IM, Price JS (2006) Evidence that the canonical Wnt signalling pathway regulates deer antler regeneration. Dev Dyn 235:1390–1399
Price JS, Oyajobi BO, Nalin AM, Frazer A, Russell RGG, Sandell LJ (1996) Chondrogenesis in the regenerating antler tip in red deer: Expression of collagen types I, IIA, IIB, and X demonstrated by in situ nucleic acid hybridization and immunocytochemistry. Dev Dyn 205:332–347
Rokutanda S, Fujita T, Kanatani N, Yoshida CA, Komori H, Liu W, Mizuno A, Komori T (2009) Akt regulates skeletal development through GSK3, mTOR, and FoxOs. Dev Biol 328:78–93
Shen YY, Lim BK, Liu HQ, Liu J, Irwin DM, Zhang YP (2012) Multiple episodes of convergence in genes of the dim light vision pathway in bats. PLoS One 7(4):e34564. https://doi.org/10.1371/journal.pone.0034564
Shin KH, Yun-Choi HS, Lim SS, Won DH, Kim JK (1999) Immuno-stimulating, anti-stress and anti-thrombotic effects of unossified velvet antlers. Nat Prod Sci 5:54–59
Steger V, Molnar A, Borsy A, Gyurjan I, Szabolcsi Z, Dancs G, Molnar J, Papp P, Nagy J, Puskas L, Barta E, Zomborszky Z, Horn P, Podani J, Semsey S, Lakatos P, Orosz L (2010) Antler development and coupled osteoporosis in the skeleton of red deer Cervus elaphus: expression dynamics for regulatory and effector genes. Mol Genet Genomics 284:273–287
Sui ZG, Zhang LH, Huo YS, Zhang YK (2014) Bioactive components of velvet antlers and their pharmacological properties. J Pharm Biomed Anal 87:229–240
Tseng SH, Sung CH, Chen LG, Lai YJ, Chang WS, Sung HC, Wang CC (2014) Comparison of chemical compositions and osteoprotective effects of different sections of velvet antler. J Ethnopharmacol 151:352–360
Vlad D, Kierzkowski D, Rast MI, Vuolo F, Dello Ioio R, Galinha C, Gan XC, Hajheidari M, Hay A, Smith RS, Huijser P, Bailey CD, Tsiantis M (2014) Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343:780–783
Wang T, Xu Z (2010) miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem Biophys Res Commun 402:186–189
Wu FF, Li HQ, Jin LJ, Li XY, Ma YS, You JS, Li SY, Xu YP (2013) Deer antler base as a traditional Chinese medicine: a review of its traditional uses, chemistry and pharmacology. J Ethnopharmacol 145:403–415
Wu X, Li S, Xue P, Li Y (2017) Liraglutide, a glucagon-like peptide-1 receptor agonist, facilitates osteogenic proliferation and differentiation in MC3T3-E1 cells through phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT), extracellular signal-related kinase (ERK)1/2, and cAMP/protein kinase A (PKA) signaling pathways involving β-catenin. Exp Cell Res 360:281–291
Wu X, Zheng S, Ye Y, Wu Y, Lin K, Su J (2018) Enhanced osteogenic differentiation and bone regeneration of poly(lactic-co-glycolic acid) by graphene via activation of PI3K/Akt/GSK-3beta/beta-catenin signal circuit. Biomater Sci 6(5):1147–1158
Yao B, Zhao Y, Wang Q, Zhang M, Liu M, Liu H, Li J (2012a) De novo characterization of the antler tip of Chinese Sika deer transcriptome and analysis of gene expression related to rapid growth. Mol Cell Biochem 364:93–100
Yao BJ, Zhao Y, Zhang HS, Zhang M, Liu MC, Liu HL, Li J (2012b) Sequencing and de novo analysis of the Chinese Sika deer antler-tip transcriptome during the ossification stage using Illumina RNA-Seq technology. Biotechnol Lett 34:813–822
Yuan Z, Liu E, Liu Z, Kijas JW, Zhu C, Hu S, Ma X, Zhang L, Du L, Wang H, Wei C (2017) Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep. Anim Genet 48:55–66
Zha EH, Gao SY, Pi YZ, Li XX, Wang YT, Yue XQ (2012) Wound healing by a 3.2 kDa recombinant polypeptide from velvet antler of Cervus nippon Temminck. Biotechnol Lett 34:789–793
Zha EH, Li XX, Li DD, Guo XS, Gao SY, Yue XQ (2013) Immunomodulatory effects of a 3.2 kDa polypeptide from velvet antler of Cervus nippon Temminck. Int Immunopharmacol 16:210–213
Zhang YC, Yu Y, Wang CY, Li ZY, Liu Q, Xu J, Liao JY, Wang XJ, Qu LH, Chen F, Xin PY, Yan CY, Chu JF, Li HQ, Chen YQ (2013) Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol 31(9):848–852
Zhang LC, Liang J, Luo WZ, Liu X, Yan H, Zhao KB, Shi HB, Zhang YB, Wang LG, Wang LX (2014) Genome-wide scan reveals LEMD3 and WIF1 on SSC5 as the candidates for porcine ear size. PLoS One 9(7):e102085. https://doi.org/10.1371/journal.pone.0102085
Zhao Y, Yao BJ, Zhang M, Wang SM, Zhang H, Xiao W (2013) Comparative analysis of differentially expressed genes in Sika deer antler at different stages. Mol Biol Rep 40:1665–1676
Zheng J, Xue H, Wang T, Jiang Y, Liu B, Li J, Liu Y, Wang W, Zhang B, Sun M (2011) miR-21 downregulates the tumor suppressor P12 CDK2AP1 and stimulates cell proliferation and invasion. J Cell Biochem 112:872–880
Acknowledgements
Thanks are due to Changchun Dongda deer Industry Co., Ltd. for its support in sample collection. This study was funded by the Agricultural Science and Technology Innovation Program of China [grant number CAAS-ASTIP-201X-ISAPS]; and the Special Economic Animals Sharing Platform in China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Pengfei Hu declares that he/she has no conflict of interest. Tianjiao Wang declares that he/she has no conflict of interest. Huamiao Liu declares that he/she has no conflict of interest. Jiaping Xu declares that he/she has no conflict of interest. Lei Wang declares that he/she has no conflict of interest. Pei Zhao declares that he/she has no conflict of interest. Xiumei Xing declares that he/she has no conflict of interest.
Ethical approval
All procedures concerning animals were organized to accord with the guidelines of care and use of experimental animals established by the Ministry of Agriculture of China, and all protocols were approved by the Institutional Animal Care and Use Committee of Institute of Special Economic Animal and Plant Sciences, Chinese Academy of Agricultural Sciences, Changchun, China.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Category of annotated genes and microRNAs. A, The overall length distributions of small RNA in heavy and light velvet antler group; B, The species distribution map of annotated genes; C, The species distribution map of annotated known microRNAs (TIF 739 KB)
Figure
S2 Statistics of the enriched KEGG categories for the differentially expressed genes. The differentially expressed genes were grouped into 12 signal pathways, among them, PI3K-Akt-signaling pathway was most relevant to velvet antler development (TIF 654 KB)
Figure S3
Validation of sequencing results by quantitative real-time RT-PCR. Transcript levels of 12 genes and 4 microRNAs, of which involve in animal organ development, in qRT-PCR analysis. The y-axis shows the relative gene expression levels analyzed by qRT-PCR. The bars represent SE (n=3). L group, light velvet antler group; H group, heavy velvet antler group (TIF 898 KB)
Rights and permissions
About this article
Cite this article
Hu, P., Wang, T., Liu, H. et al. Full-length transcriptome and microRNA sequencing reveal the specific gene-regulation network of velvet antler in sika deer with extremely different velvet antler weight. Mol Genet Genomics 294, 431–443 (2019). https://doi.org/10.1007/s00438-018-1520-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-018-1520-8