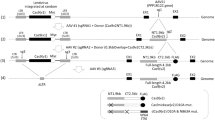
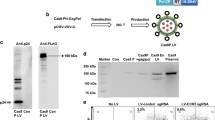
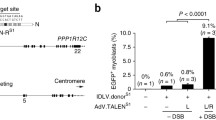
Abstract
Targeted integration of transgenes facilitates functional genomic research and holds prospect for gene therapy. The established microhomology-mediated end-joining (MMEJ)-based strategy leads to the precise gene knock-in with easily constructed donor, yet the limited efficiency remains to be further improved. Here, we show that single-strand DNA (ssDNA) donor contributes to efficient increase of knock-in efficiency and establishes a method to achieve the intracellular linearization of long ssDNA donor. We identified that the CRISPR/Cas9 system is responsible for breaking double-strand DNA (dsDNA) of palindromic structure in inverted terminal repeats (ITRs) region of recombinant adeno-associated virus (AAV), leading to the inhibition of viral second-strand DNA synthesis. Combing Cas9 plasmids targeting genome and ITR with AAV donor delivery, the precise knock-in of gene cassette was achieved, with 13–14% of the donor insertion events being mediated by MMEJ in HEK 293T cells. This study describes a novel method to integrate large single-strand transgene cassettes into the genomes, increasing knock-in efficiency by 13.6–19.5-fold relative to conventional AAV-mediated method. It also provides a comprehensive solution to the challenges of complicated production and difficult delivery with large exogenous fragments.





Similar content being viewed by others
Abbreviations
- CRISPR/Cas9:
-
Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein-9 nuclease
- MMEJ:
-
Microhomology-mediated end-joining
- ssDNA:
-
Single-strand DNA
- dsDNA:
-
Double-strand DNA
- ITR:
-
Inverted terminal repeat
- AAV:
-
Adeno-associated virus
- gRNA:
-
Guide RNA
- DSB:
-
Double-strand break
- HDR:
-
Homology-directed repair
- NHEJ:
-
Non-homologous end joining
- HA:
-
Homology arm
- PITCH:
-
Precise integration into target chromosome
- ssODN:
-
Single-strand oligodeoxynucleotide
- HR:
-
Homologous recombination
- HP:
-
Hairpin
- PAM:
-
Protospacer adjacent motif
- SNP:
-
Single nucleotide polymorphism
- RNP:
-
Ribonucleoprotein
- HEK:
-
Human embryonic kidney
- PEI:
-
Polyethylenimine
- qRT-PCR:
-
Quantitative real-time PCR
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- FBS:
-
Fetal bovine serum
References
Aida T, Nakade S, Sakuma T, Izu Y, Oishi A, Mochida K, Ishikubo H, Usami T, Aizawa H, Yamamoto T, Tanaka K (2016) Gene cassette knock-in in mammalian cells and zygotes by enhanced MMEJ. BMC Genom 17:979
Armstrong GA, Liao M, You Z, Lissouba A, Chen BE, Drapeau P (2016) Homology directed knockin of point mutations in the zebrafish tardbp and fus genes in ALS using the CRISPR/Cas9 system. PLoS One 11:e0150188
Asuri P, Bartel MA, Vazin T, Jang JH, Wong TB, Schaffer DV (2012) Directed evolution of adeno-associated virus for enhanced gene delivery and gene targeting in human pluripotent stem cells. Mol Ther 20:329–338
Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD (2011) High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods 8:753–755
Gaj T, Staahl BT, Rodrigues GMC, Limsirichai P, Ekman FK, Doudna JA, Schaffer DV (2017) Targeted gene knock-in by homology-directed genome editing using Cas9 ribonucleoprotein and AAV donor delivery. Nucleic Acids Res 45:e98
Galli A, Cervelli T (2014) Inverted terminal repeats of adeno-associated virus decrease random integration of a gene targeting fragment in Saccharomyces cerevisiae. BMC Mol Biol 15:5
Hirata R, Chamberlain J, Dong R, Russell DW (2002) Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat Biotechnol 20:735–738
Huai C, Jia C, Sun R, Xu P, Min T, Wang Q, Zheng C, Chen H, Lu D (2017) CRISPR/Cas9-mediated somatic and germline gene correction to restore hemostasis in hemophilia B mice. Hum Genet 136:875–883
Khan IF, Hirata RK, Russell DW (2011) AAV-mediated gene targeting methods for human cells. Nat Protoc 6:482–501
Kim S, Kim D, Cho SW, Kim J, Kim JS (2014) Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24:1012–1019
Lin S, Staahl BT, Alla RK, Doudna JA (2014) Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 3:e04766
Ling C, Wang Y, Lu Y, Wang L, Jayandharan GR, Aslanidi GV, Li B, Cheng B, Ma W, Lentz T, Ling C, Xiao X, Samulski RJ, Muzyczka N, Srivastava A (2015) Enhanced transgene expression from recombinant single-stranded D-sequence-substituted adeno-associated virus vectors in human cell lines in vitro and in murine hepatocytes in vivo. J Virol 89:952–961
Mao Z, Bozzella M, Seluanov A, Gorbunova V (2008) DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 7:2902–2906
Maresca M, Lin VG, Guo N, Yang Y (2013) Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res 23:539–546
Miller DG (2011) AAV-mediated gene targeting. Methods Mol Biol 807:301–315
Nakade S, Tsubota T, Sakane Y, Kume S, Sakamoto N, Obara M, Daimon T, Sezutsu H, Yamamoto T, Sakuma T, Suzuki KT (2014) Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun 5:5560
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F (2014) CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159:440–455
Quadros RM, Miura H, Harms DW, Akatsuka H, Sato T, Aida T, Redder R, Richardson GP, Inagaki Y, Sakai D, Buckley SM, Seshacharyulu P, Batra SK, Behlke MA, Zeiner SA, Jacobi AM, Izu Y, Thoreson WB, Urness LD, Mansour SL, Ohtsuka M, Gurumurthy CB (2017) Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol 18:92
Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520:186–191
Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE (2016) Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34:339–344
Russell DW, Hirata RK (1998) Human gene targeting by viral vectors. Nat Genet 18:325–330
Sakuma T, Takenaga M, Kawabe Y, Nakamura T, Kamihira M, Yamamoto T (2015) Homologous recombination-independent large gene cassette knock-in in CHO cells using TALEN and MMEJ-directed donor plasmids. Int J Mol Sci 16:23849–23866
Sakuma T, Nakade S, Sakane Y, Suzuki KT, Yamamoto T (2016) MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc 11:118–133
Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, Haliburton GE, Ye CJ, Bluestone JA, Doudna JA, Marson A (2015) Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci USA 112:10437–10442
Siu JJ, Queen NJ, Huang W, Yin FQ, Liu X, Wang C, McTigue DM, Cao L (2017) Improved gene delivery to adult mouse spinal cord through the use of engineered hybrid adeno-associated viral serotypes. Gene Ther 24:361–369
Wu Z, Yang H, Colosi P (2010) Effect of genome size on AAV vector packaging. Mol Ther 18:80–86
Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, Aach J, Kim-Kiselak C, Briggs AW, Rios X, Huang PY, Daley G, Church G (2013) Optimization of scarless human stem cell genome editing. Nucleic Acids Res 41:9049–9061
Yao X, Wang X, Hu X, Liu Z, Liu J, Zhou H, Shen X, Wei Y, Huang Z, Ying W, Wang Y, Nie YH, Zhang CC, Li S, Cheng L, Wang Q, Wu Y, Huang P, Sun Q, Shi L, Yang H (2017a) Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res 27:801–814
Yao X, Wang X, Liu J, Hu X, Shi L, Shen X, Ying W, Sun X, Wang X, Huang P, Yang H (2017b) CRISPR/Cas9—mediated precise targeted integration in vivo using a double cut donor with short homology arms. EBioMedicine 20:19–26
Acknowledgements
We are grateful to Prof. Hui Yang for kindness gift of Cas9 expressing vector and MMEJ plasmid, to Dr. Cong Huai and Mr. Chenqiang Jia for their help in language polishing, to Mr. Hairui Xi and Mr. Renji Zhang for providing technical assistance.
Funding
This study was funded by the National Natural Science Foundation of China (Grant Nos. 81772657, 31571371 and 81372706) and Precision Medicine Research Key Project of National Key R&D Plan (Grant No. 2017YFC0907501).
Author information
Authors and Affiliations
Contributions
HC and DL conceptualized and designed the project. QX and TM performed all the experiments. QX and LH analyzed and interpreted the data. QX and SM wrote the manuscript, which all authors edited and approved.
Corresponding authors
Ethics declarations
Conflict of interest
Qing Xiao declares that she has no conflict of interest. Taishan Min declares that she has no conflict of interest. Shuangping Ma declares that she has no conflict of interest. Lingna Hu declares that she has no conflict of interest. Hongyan Chen declares that she has no conflict of interest. Daru Lu declares that he has no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Additional information
Communicated by S. Hohmann.
Hongyan Chen and Daru Lu are co-corresponding authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xiao, Q., Min, T., Ma, S. et al. Intracellular generation of single-strand template increases the knock-in efficiency by combining CRISPR/Cas9 with AAV. Mol Genet Genomics 293, 1051–1060 (2018). https://doi.org/10.1007/s00438-018-1437-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-018-1437-2