Abstract
Ascorbate peroxidase (APX) is a member of heme-containing peroxidases which catalyze the H2O2-dependent oxidation of a wide range of substrates in plants and animals. As is known, H2O2 acts as a signaling molecule in the regulation of fiber development. Our previous work reported that ascorbate peroxidase 1 (GhAPX1) was important for cotton fiber elongation. However, knowledge about APX gene family members and their evolutionary and functional characteristics in cotton is limited. Here, we report 26 GhAPX genes by genome-wide investigation of tetraploid cotton Gossypium hirsutum. Phylogenetic and gene structure analyses classified these APX members into five clades and syntenic analysis suggested two duplication events. Expression profiling of the 26 APXs revealed that ten members are expressed in cotton fibers. Notably, GhAPX10A, GhAPX10D, GhAPX12A, and GhAPX12D showed high expression levels in 30-day fiber, while GhAPX1A/D, GhAPX3A/D, and GhAPX6A/D showed very low expression levels. The enzyme activity and H2O2 content assays revealed that cotton fiber kept high enzyme activity and the lowest H2O2 level in 30-day fibers, indicating that other than GhAPX1, the newly reported APX members are responsible for the reactive oxygen species homeostasis in the cotton fiber maturation stages. Expression profiling of ten fiber-expressed APXs after phytohormone treatments revealed their regulation patterns by different stimuli, suggesting that GhAPX1, GhAPX12A, and GhAPX12D are responsible to most phytohormone treatments. Our data provided evolutionary and functional information of GhAPX gene family members and revealed that different members are responsible to redox homeostasis during different cotton fiber development stages.
Similar content being viewed by others
Introduction
Tetraploid upland cotton (Gossypium hirsutum) is the most widely cultivated cotton plants, which is the most important resource of natural fiber for textile industry (Zhu 2016). The genome of tetraploid cotton (G. hirsutum, AADD, 2n = 4x = 52) and its diploid ancestor cotton (Gossypium arboretum, AA, 2n = 2x = 26; Gossypium raimondii, DD, 2n = 2x = 26) had been sequenced recent years (Paterson et al. 2012; Wang et al. 2012; Li et al. 2014, 2015; Zhang et al. 2015). Benefit from the publication of the genome of upland cotton (70,478 predicted protein coding genes), a large number of functional genes and their gene family members involved in cotton fiber development have been reported (Huang et al. 2015; Guo et al. 2016; Zhang et al. 2016; Wu et al. 2017). However, in-depth digging of the genome data will be necessary.
Reactive oxygen species (ROS) can be continuously produced in all aerobic organisms to take important role as regulator for cellular response to environmental factors in plants (Alscher et al. 1997; Pandey et al. 2017). It is demonstrated that H2O2, a signaling molecule known as the major active ROS type, is involved in regulation of plant cell development and stress resistance, such as root hair initiation and elongation, drought and salinity resistance, and temperature stress response (Pei et al. 2000; Panchuk et al. 2005; Pinheiro and Chaves 2011; Qu et al. 2013). ROS could synergize or antagonize many cellular regulatory circuits through active interaction with other signals and plant hormones during growth, development, and stress responses (Petrov and Breusegem 2012; Tao et al. 2016). In plant cells, despite the vital role in cell development, ROS accumulation could also cause severe damages, accordingly, plants developed a complex antioxidant system to prevent cellular damage generated by ROS, such as ascorbic acid (AsA), glutathione (GSH), and carotenoids (Smirnoff 2000).
Ascorbate peroxidase (APX, EC 1.11.1.11) is a family of type I heme-containing peroxidase that catalyzes H2O2 to water using ascorbate as specific electron donor, functioning in maintaining cell reduction/oxidation (redox) homeostasis by scavenging ROS (Foyer and Halliwell 1976; Noctor and Foyer 1998; Sharp et al. 2003; Suzuki et al. 2012). APXs are encoded by small multigene families in higher plants and are classified into different groups according to their subcellular localization (Teixeira et al. 2004). In Arabidopsis, eight APXs have been identified with localization of three in cytosol, three in peroxisome, and two in chloroplast, respectively (Shigeoka et al. 2002; Chew et al. 2003). APX family has been fully characterized in rice and tomato based on their genomes (Teixeira et al. 2006; Najami et al. 2008).
Cotton fiber, composed of numerous non-branched single cell, is an ideal model material to investigate cell growth (Li et al. 2017). ROS promoted cell expansion or enlargement though participating in plant cell wall loosening (Cosgrove 2000; Liszkay et al. 2004). It has been demonstrated that ROS plays vital role in cotton fiber cell elongation development (Li et al. 2007; Qin et al. 2008; Mei et al. 2009). Previously, we reported that GhAPX1 plays a significant role in cotton fiber elongation via involving in ethylene signaling pathway (Li et al. 2007). Over-expression of cotton GhAPX1A/D increased fiber resistance to H2O2 stress (Guo et al. 2016). However, detailed knowledge about the whole APX family and expression patterns of APX genes in G. hirsutum remains unclear.
In this work, we performed genome-wide investigation and expression profiling of APX family in G. hirsutum. A total of 26 GhAPX genes were identified. Phylogenetic and gene structure analyses classified these APX members into five clades and syntenic analysis suggested two duplication events. Expression profiling of the 26 APXs revealed that ten members are expressed in cotton fibers. Interestingly, GhAPX10A, GhAPX10D, GhAPX12A, and GhAPX12D showed high expression levels in 30-day fiber, while GhAPX1A/D, GhAPX3A/D, and GhAPX6A/D showed very low expression levels at the same development stage. The enzyme activity and H2O2 content assays revealed that cotton fiber kept high enzyme activity and the lowest H2O2 level in 30-day fibers, indicating that other than GhAPX1, the newly reported APX members are responsible for the reactive oxygen species homeostasis in the secondary cell wall biosynthesis and maturation of cotton fiber development stages. This work provided evolutionary and functional information of GhAPX gene family members and revealed that different GhAPX family members are responsible to redox homeostasis during different cotton fiber development stages.
Materials and methods
Plant growth and different treatments
Cotton plants (G. hirsutum L. cv. Xuzhou 142) and the fuzzless-lintless mutant (fl) were grown in an experimental field at the Institute of Tropical Biosciences and Biotechnology in Haikou, China. Cotton bolls were labeled on the day of anthesis (defined as 0d ) and then detached in different developmental stages (5d, 10d, 15d, 20d, 25d, and 30d). The 10d fibers were treated with 200 µm ethephon (ETH), 100 µm H2O2, 100 µm gibberellin (GA), 100 µm methyl jasmonate (MeJA), 100 µm brassinolide (BR), and 1 mg/L of indole-3-acetic acid (IAA) for 1, 3, 6, and 12 h, respectively (Xin et al. 2016). Different tissues of roots, stems, leaves, petals, anthers, ovules, and fibers were immediately frozen in liquid nitrogen, and then stored at − 80 °C after stripped and separated from each other.
Identification and multiple alignment of GhAPXs
Genome data of G. hirsutum were downloaded from Cotton Genome Project (http://cgp.genomics.org.cn/page/species/index.jsp) and CottonGen database (https://www.cottongen.org/) as described (Li et al. 2015; Zhang et al. 2015). The APX sequences of Arabidopsis, Oryza sativa, and Theobroma cacao were used as seed sequences to obtain the cotton APXs by local BLASTP through searching with a cutoff e value of 1e−10. Obtained 26 GhAPXs were submitted to InterProScan (http://www.ebi.ac.uk/interpro/) to assess the APX domains (IPR002016). Multiple sequence alignment was performed using ClustalX (2.0) with default parameter (Larkin et al. 2007).
Chromosomal location analysis and phylogenetic tree construction
MapInspect software was used to visualize the distribution of the 26 APX genes in G. hirsutum chromosomes. Phylogenetic tree was constructed using MEGA 5.1 software with neighbor-joining method and bootstrap values of 1000 replicates as described (Tamura et al. 2007).
Intron–exon and motif structure analysis of APX family
Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/index.php) was used to analyze the intron–exon structure by comparing the CDS of APX genes with their corresponding genomic sequences (Hu et al. 2015). Deduced protein sequences of GhAPXs were submitted to multiple expectation maximization for motif elicitation (MEME) program for the identification of the conserved motifs (Bailey et al. 2006).
Syntenic and evolutionary analyses
Paralogous GhAPX gene pairs were estimated based on their nucleotide identities > 90%. Tandem duplication events occurred when two closely related GhAPX genes are located within the same chromosome region. Segmental duplication has been defined as paralogous genes. The syntenic relationships of paralogous and orthologous between cotton and a closely related cacao species were analyzed using Circos program (Krzywinski et al. 2009) based on sequence identity calculations and the phylogenetic tree.
Evolutionary analyses were performed as previously reported (Jin et al. 2017). Briefly, the Ka (nonsynonymous substitution rate) and Ks (synonymous substitution rate) were calculated by DnaSP 5.0 software. The Ka/Ks ratios for GhAPX genes were used to assess the selection pressure on duplicated genes and Ka/Ks ratio > 1, < 1, or = 1 indicates positive, negative, or neutral evolution, respectively. Furthermore, Tajima relative rate tests were performed to determine the equality of the evolutionary rate between GhAPX paralogues and orthologues.
RNA extraction and qRT-PCR
Total RNA was extracted from different cotton tissues by a modified hot borate method as described (Shi et al. 2006). Five micrograms of total RNA for each tissue were used to synthesize first-strand cDNA using SuperScript® III first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA, USA). Reverse transcript PCR (RT-PCR) and quantitative real-time PCR (qRT-PCR) were performed using the SYBR green real-time PCR master mixes (Appliedbiosystems, Foster, CA, USA) with specific primers provided in Table S1. The 5′- and 3′-UTR of GhAPXs were obtained by genome-referenced expressed sequence tags (ESTs) assembly as described to facilitate the gene-specific primer design (Jin et al. 2013). The UBQ gene was used as internal control to adjust the amount of template cDNA for quantitative analysis (Jin et al. 2016). The relative expression level of each APX gene was used to generate a heat map using MultiExperiment viewer (MeV, version 4.9) software.
Cis-regulatory elements analysis
The promoters of GhAPXs were downloaded in local database and the Plant CARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to analyze the cis-regulatory element of GhAPXs promoter (Lescot et al. 2002).
Determination of APX enzyme activity and H2O2 content
Different fiber tissues (5d, 10d, 15d, 20d, 25d, and 30d) were used to measure the APX enzyme activity and H2O2 content as described (Li et al. 2007).
Results
Identification of the APX gene family in G. hirsutum
Several reports indicated that GhAPX1 plays important roles during cotton fiber initiation and elongation stages (Fig. S1) (Shi et al. 2006; Li et al. 2007; Yang et al. 2008; Zheng et al. 2014; Guo et al. 2016). To further understand the functional of APX gene family, a genome-wide investigation of GhAPXs was performed. A total of 26 non-redundant GhAPX genes (Table S2) were identified by searching the cotton genome database, and were renamed from GhAPX1A to GhAPX13A and from GhAPX1D to GhAPX13D according to their order in chromosomes of the A and D sub-genomes except for previously reported GhAPX1A and GhAPX1D. The detailed information of these genes were listed in Table 1, including chromosome location, ORF length, protein length, molecular weight, and theoretical isoelectric point.
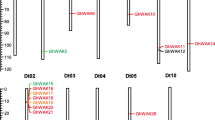
Chromosome distribution analysis showed that GhAPX1A–GhAPX5A, GhAPX7A, GhAPX8A, and GhAPX10A–13A were anchored in eight chromosomes of A sub-genome, while GhAPX6A and GhAPX9A were anchored in two un-assembled scaffolds: A-Chr5 scaffold1211 and A-Chr6 scaffold1353, respectively (Fig. 1a, b). For D sub-genome, GhAPX2D–GhAPX13D were anchored in eight chromosomes, and GhAPX1D was anchored in D-Chr5 scaffold4074 (Fig. 1c, d). The 26 GhAPXs were predicted to be located in different apparatus including 18 (9 ortholog pairs) in cytoplasm, four in periplasm, and four in outer membrane (Table S3).
Chromosome distribution of GhAPX gene family. The 26 GhAPX genes were mapped to the chromosomes of A sub-genome (a) and un-assembled scaffolds (b), as well as that of D sub-genome (c) and un-assembled scaffold (d). Genes were referred as GhAPX1A–13A and GhAPX1D–13D, according to their organization order on the chromosomes, except for the previously reported GhAPX1A/D. Different scales were used for chromosomes and un-assembled scaffolds. Note that the chromosome information of scaffolds was known, although they could not be assembled to the corresponding chromosomes
Phylogenetic and intron–exon distribution analyses of GhAPX gene family
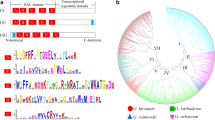
To in-depth understand the evolutionary and phylogenetic relationships of GhAPXs, a neighbor-joining (NJ) phylogenetic tree was constructed using protein sequences of APXs from Arabidopsis thaliana, G. hirsutum, O. sativa, Vitis vinifera, and T. cacao (Fig. 2a). The 26 GhAPXs could be classified into five well-supported clades labeled with different colours. According to A. thaliana APXs (Panchuk et al. 2005), clade I and clade II consist of cytoplasmic APXs, Clade III contains all known chloroplast APXs, and Clade IV and V include peroxisomal APXs. These phylogenetic data demonstrated similar conclusions to a previously published work (Guo et al. 2016).
Phylogenetic and intron–exon structure analyses of APX family genes. a Protein sequences of APX gene families from Arabidopsis thaliana (AtAPX), G. hirsutum (GhAPX), Oryza sativa (OsAPX), Theobroma cacao (TcAPX), and Vitis vinifera (VvAPX) were used to construct an NJ phylogenetic tree. Bootstrap was set to 1000 replicates. Five clades were named as sub-family I to V, shadowed with different colours. b Intron–exon organization structure analysis of the 26 GhAPX genes was shown. Boxes and lines represented the exons and introns, and the genomic length was indicated at the bottom. (Color figure online)
Gene structures of all the 26 GhAPXs were investigated to further validate the evolution and phylogenetic relationships of GhAPX family members. The GhAPX genes belonged to the same clade in phylogenetic tree shared similar intron–exon organization structures (Fig. 2b).
Syntenic and evolutionary analyses of GhAPX gene family
To investigate the expansion of the APX gene family, syntenic analysis of G. hirsutum and T. cacao APXs was performed using Circos software. Two tandem duplication events were detected in both A- and D-sub-genome (GhAPX10A/11A/12A and GhAPX10D/11D/12D); however, no segmental duplication events were determined because of the high conservation of GhAPXs between A- and D-sub-genome (Fig. 3).
Tandem duplication and segmental duplication of GhAPXs and syntenic analysis between G. hirsutum and T. cacao APXs. Chromosomes and scaffolds from cotton (Gh) and cacao (Tc) are shown in yellow and blue segments, respectively. The positions of the APX genes are marked by black lines in the circus. Duplicated GhAPXs are linked by red lines and syntenic relationships between cotton and cacao are linked by purple lines. (Color figure online)
Evolutionary selection patterns between paralogue and/or orthologue gene pairs can be estimated by the Ka/Ks ratio (Yadav et al. 2015). A Ka/Ks ratio > 1 indicates a positive selection, a Ka/Ks ratio < 1 indicates a purifying selection, and a Ka/Ks ratio = 1 indicates a neutral selection. The Ka/Ks ratios of the duplicated GhAPXs indicated that they all were subjected to purifying selection (Table 2). In addition, Tajima relative rate were calculated to determine whether the GhAPX duplicates evolved at an accelerated rate following the duplication events. Notably, statistically significant increase in evolutionary rate occurred between the GhAPX10A/11A/12A duplicated paralogues, while non-significant evolutionary rate occurred between GhAPX10D/11D/12D (Table 3), indicating a potential functional divergence of these duplicated paralogues.
Conserved motif analysis of GhAPX family
Multiple sequence alignment showed that all GhAPXs had three conserved domains and variable N-terminus (Fig. S2). Domain I contains two active sites, Domain II has the most conserved 12 amino acids sequences that predicted as Heme-binding site, and Domain III has three proximal cation-binding sites. These data provide possibility that GhAPXs may function in different organelles using ascorbate as substrate to detoxify H2O2.
Conserved motifs in GhAPXs were searched by MEME program to obtain more insights into the diversity of motif compositions and evolutionary relationships, and a total of ten conserved motifs were discovered. The APXs belong to the same clades that share very similar motif composition and order (Fig. 4a). Motif 1, 2, and 7 are existed in all GhAPXs, indicating that they are conserved sections of GhAPXs. Most of cytoplasmic GhAPXs have motifs 1–8, except for orthologs GhAPX4A and GhAPX4D. Chloroplast GhAPXs have all motifs but not motif 6, while the peroxisomal APXs have the least conserved motifs 1, 2, 7, and 9. Motif 10 is only distributed in the chloroplast-located APXs, with a most conserved amino acid sequence of GWGKPETKYTKDGPG (Fig. 4a, b). Motif 9 is observed in the C-terminal of cytoplasmic GhAPX3A/D and GhAPX5A, while in the N-terminal of the peroxisome- and chloroplast-located GhAPXs.
Motif analysis of GhAPXs. Conserved motifs of GhAPX protein sequences were analyzed (a). Ten different motifs were recognized and indicated with different colours. The organization order of motifs for each member of GhAPXs was highlighted. The conservation of the sequences for each conserved domain was also presented (b). (Color figure online)
Tissue- and development-specific expression profiling of GhAPX genes
To understand the expression and function diversity of the 26 GhAPX genes, the tissue- and development-specific expression profiles of GhAPXs were performed using qRT-PCR. Relative expression levels of the 26 GhAPX genes in eight different tissues were demonstrated to construct a heat map (Fig. 5a). All expression level data were normalized using cotton UBQ as internal control, and relative expression level over 0.05-fold to UBQ was considered to be detected. Members of cytoplasmic GhAPX sub-family I were universal expressed, in which GhAPX1A/D, GhAPX10A, GhAPX10D, GhAPX12A, and GhAPX12D were the predominantly expressed GhAPXs, with high levels in leaf, petal, and anther. Members of cytoplasmic GhAPX sub-family II were expressed in much less tissues: GhAPX5A/D was expressed only in leaf, and GhAPX3A/D was expressed in leaf, petal, and anther, indicating the functional diversity of the two clusters of GhAPXs. However, none of chloroplast GhAPXs were detected, except for GhAPX6A/D. For peroxisomal GhAPX sub-family IV and V, GhAPX7A was expressed in leaf and anther, while GhAPX7D expressed only in leaf. The semi-quantitative RT-PCR results were also showed with visible confirmation (Fig. 5b).
Expression profiling of GhAPX genes in different cotton tissues and different development stage of cotton fibers. a Heat map of the qRT-PCR data for the 26 GhAPXs in eight different cotton tissues. Orthologs with very high identities that could not be distinguished by gene-specific primers were examined together use the same primers and indicated by slashes. b Semi-quantitative RT-PCR results of GhAPX genes corresponding to that of in a. c Heat map of the qRT-PCR data for ten fiber-preferentially accumulated GhAPXs in eight development stages of cotton fibers. Orthologs GhAPX1A/D, GhAPX3A/D, and GhAPX6A/D were detected use the same primers, respectively. d Semi-quantitative RT-PCR results of GhAPX genes corresponding to that of in c. Relative expression levels were normalized by the internal control UBQ gene
Interestingly, ten GhAPX genes were detected in 10-day fibers (Fiber-10), and selected to further examine the expression patterns during cotton fiber development. Fibers of eight developmental stages were used for qRT- and RT-PCR analyses, including 5-day (Fiber-5), 10-day (Fiber-10), 15-day (Fiber-15), 20-day (Fiber-20), 25-day (Fiber-25), 30-day (Fiber-30), 10-day WT ovule (Ovule-10), and 10-day fl ovule (fl-Ovule-10). The results showed that these ten GhAPX genes displayed three distinct expression patterns according to different temporal expression feature during fiber development stages. GhAPX1A/D had a predominant steady expression level from 5 days to 25 days of fiber elongation and secondary cell wall biosynthesis. Notably, GhAPX6A/D and GhAPX3A/D were mainly expressed at the late fiber development stage of secondary cell wall biosynthesis (Fig. 5c, d). Meanwhile, GhAPX10A, GhAPX10D, GhAPX12A, and GhAPX12D had the highest expression level at 30 days, the cell apoptosis stage of fiber development. The results of the 26 GhAPX gene expression patterns provide their probable multiple functions in cotton plant development, particularly the potential diverse role in controlling H2O2 concentration during different fiber development stages.
Determination of APX activity and H2O2 content during fiber development
To understand the relationship between APX expression and H2O22homeostasis during fiber development, different tissues of 5d, 10d, 15d, 20d, 25d, and 30d fibers were collected to measure the APX activity and H2O2 content. The level of APX activity reached the peak value in 5d fibers, and maintained a steady high expression with a tendency of decline at fast fiber elongation stages (5–15 dpa), following a slight increase at 20 dpa, which is matched well to the gene expression level of GhAPXs, indicating that there may be diverse GhAPXs functioning in different developmental stages (Fig. 6a). Meanwhile, H2O2 content demonstrated an ascending trend at the fast fiber elongation stages (5–15 dpa) with highest concentration in 20d fibers, and then decrease gradually at the secondary cell wall synthesis stages (20–30 dpa). The results imply the possibility that some GhAPXs accumulated in the secondary cell wall biosynthesis and maturation stages of fiber development may involve in H2O2 scavenging (Fig. 6b).
Analyses of cis-regulatory element and gene expression profiling of GhAPXs in response to stimulations of H2O2 and phytohormone
To further investigate the regulatory mechanism of the GhAPX gene family members, especially the duplicated paralogues, the cis-elements were scanned in the promoter regions of GhAPXs (Fig. 7a–e). A 1500-bp sequence upstream of the translational start site was considered as a putative promoter region, and thus was used to analyze the distribution of cis-regulatory elements. The cis-elements were characterized and indicated with capital letters labeled by different colours, including two core cis-elements, nine stress response elements, and six phytohormone response elements which were characterized (Fig. 7, Table S4), which implies that the GhAPXs gene expression is under control of stimulation responsiveness of stress and phytohormone. Notably, in the process of cotton plant growth and development, similar cis-element distribution pattern was found in the promoter regions of the duplicated paralogues of common or higher expressing GhAPX genes, while the duplicated paralogues of lower or non-expressing GhAPX genes displayed different distributions with non-regular constitutions.
Cis-regulatory elements analysis and expression profiling of GhAPXs response to plant hormone (a–e). The putative cis-regulatory elements of GhAPXs were labeled with capital letters in the figure according to their relative position. The green letters stand for stress response cis-elements, the blue letters stand for plant hormone response cis-elements, and the red letters stand for transcription initiation cis-elements. More details about cis-regulatory elements are shown in S4 Table. f Heat map of the qRT-PCR data for ten fiber-preferentially accumulated GhAPXs in 10-day fibers treated with different plant hormones at different hours. Orthologs GhAPX1A/D, GhAPX3A/D, and GhAPX6A/D were detected using the same primers, respectively. Relative expression levels were normalized by the internal control UBQ gene. (Color figure online)
In view of H2O2 and phytohormone important functions in cotton fiber development, PKc enzyme activity is positive related to the H2O2 content and negatively correlated with fast fiber elongation in cotton. Phytohormones such as auxins, ethylene, and brassinosteroids are involved in regulation of fiber development (Shi et al. 2006; Pang et al. 2010; Chen and Guan 2011; Zhang et al. 2011, 2016). The expression patterns of GhAPXs in cotton fibers under oxidative stress and phytohormone stimulation were determined by treating 10-day WT fibers by H2O2, ETH, IAA, BR, JA and GA for 1, 3, 6, and 12 h. The results indicated that GhAPX1A/D, GhAPX6A/D, GhAPX12A, GhAPX12D were positively respond to ethylene stimulation. GhAPX6A/D, GhAPX10A, and GhAPX12A displayed induced expression after GA treatment, while GhAPX10A, GhAPX10D, and GhAPX12A were significantly increased after JA stimulation. No changes were detected after treatments of IAA and BR. Remarkably, all the fiber-expressed GhAPX members except for GhAPX3A/D illustrated significant induced expression after H2O2 treatment (Fig. 7f). These results suggested that GhAPX genes may perform multiple functions in the process of H2O2 and phytohormone regulated cotton fiber development. In addition, there exists the appearance that duplicated paralogues displayed different responsive characteristics, implying their functional and regulatory diversity (Fig. 7f).
Discussion
DNA sequencing data of the cotton genome provide us valuable information of gene family in Gossypium to further understand gene function and regulation mechanism (Yao et al. 2012). Ascorbate peroxidase is known as the key enzyme detoxifying H2O2 and performs vital roles in plant growth and development and stress responsiveness (Fryer et al. 2003; Davletova et al. 2005). Whereas the diverse functions of GhAPX members remain unclear, especially in cotton fiber development. Thus, to comprehensively understand GhAPXs’ various roles and the regulatory mechanism, based on our previous study about GhAPX1, here, a complete overview of this GhAPX family in G. hirsutum is presented, as well as the expression profiling characteristics. Totally, 26 APX genes were identified according to the complete genome of G. hirsutum, locating onto 8 chromosomes of A- or D-sub-genome (Fig. 1; Table 1), while only 8, 8, 7, and 7 APX genes were characterized in Arabidoisis, O. sativa, T. cacao, and V. vinifera, respectively (Panchuk et al. 2005; Teixeira et al. 2006).
Phylogenetic analysis of the reported 74 APX members in different plant species showed that the APXs can be classified into four clades with different putative subcellular locations (Teixeira et al. 2004). However, the 26 cotton GhAPXs reported here were divided into five sub-families with putative different subcellular distributions according to the orthologous APX genes from A. thaliana (Fig. 2a). Intron–exon structure analysis presented high consistence with phylogenetic classification (Fig. 2b). Different APX sub-families displayed lower identities, indicating significant original and functional diversity of GhAPX gene family.
The alignment of 74 reported plant APX sequences revealed two signatures in plant chloroplast isoforms including 7 residues next to the active site (K-[ND]-I-[ETK]-E-W-P), and 16 residues near heme-binding site (E-T-K-Y-T-[KE]-[DNTE]-G-PG-[ANEK]-[PA]-G-G-Q-S), respectively. Phylogenetic analysis among different species showed that these 74 APXs were classified into 4 clades with different subcellular locations (Teixeira et al. 2004). We found that all APX proteins contain three conserved domains (Fig. S2). Study of APX gene family in O. sativa illustrated that a sequence of gene duplications led to the current diversity of isoforms (Teixeira et al. 2004), suggesting that these unique motifs may be responsible for diverse functions in different isoforms. The specific exon I and II of chloroplast APX gene SlApx7 and SlApx6 encoded for the organellar targeting sequences of the proteins (Najami et al. 2008). Under these circumstances, we deduce that the particular conserved motifs in chloroplast and cytoplasmic GhAPXs, as well as different intron–exon structures (Fig. 2 and Table S3) may have their specific possible action in targeting the organelles.
Gene expression patterns are usually closely related to their functions, and analyses of differential expression profiles can provide important information with gene families. (Guo et al. 2008). Eight APX members containing three cytosolic, two chloroplastic, and three microsomal isoforms were characterized in A. thaliana, in which APX1 and APX3 were appeared to be high expressed, while APX2 and APX5 were low expressed between different age leaves (Panchuk et al. 2005). KO-APX1 experiment in A. thaliana showed that cytosolic APX1 plays an important role in protecting chloroplast from H2O2 damage, and stromal/mitochondrial APX can be the first chloroplast line to defend against the diffusion of H2O2 from cytosol into the chloroplast (Davletova et al. 2005). In Solanum lycopersicum, dominant expressions of SlAPX6 in leaves and SlAPX7 in stems were observed (Najami et al. 2008). Members of GhAPX family were characterized with different expression profiles in root, leaf, anther, and fiber, ten GhAPX genes demonstrated fiber-specific expressions with distinct patterns according to their abundant accumulation in different stages of fiber development (Fig. 5). It has been demonstrated that GhAPX1 is highly up-regulated during fiber fast elongating stages (Li et al. 2007; Yang et al. 2008; Zheng et al. 2014; Guo et al. 2016), which is consistent with the current result that GhAPX1A/D are mainly expressed in the elongation and secondary cell wall biosynthesis stages of fiber development. Interestingly, in the secondary cell wall biosynthesis and maturation stages of fiber development, four genes of GhAPX10A/D and GhAPX12A/D were mostly enriched; meanwhile, H2O2 content indicated a tendency of decline (Fig. 6), indicating potential possibility that the four GhAPXs may be major members controlling intracellular H2O2 levels in the maturation stages of fiber development.
H2O2 and phytohormone are key factors in regulating fiber development (Li et al. 2007; Triplett et al. 2007). Many genes have been investigated that perform essential functions in fiber development through responding to phytohormone, cotton CesA were reported to increase fiber number per seed after auxin and gibberellin treatments (Triplett et al. 2007), gibberellin could induce significant expression of cotton KCS gene in cotton fibers (Xiao et al. 2016), and the transcription of the cotton AOCs was increased after JA treatment (Wang et al. 2015). Our previous work showed that GhAPX1 is involved in the response to ethylene and H2O2 stimulations (Li et al. 2007). The current investigations of the expression profiles of the APX genes in response to phytohormone and H2O2 treatments indicated that fiber-expressed GhAPXs are expressed under control of ethylene, GA, and JA. Notably, fiber-expressed GhAPXs except for GhAPX3A/D were responded to H2O2 stimulation, suggesting that these GhAPXs may be participated in redox homeostasis. Distribution analysis of cis-elements of the promoter regions of GhAPX genes supplies potential regulation mechanism of APX responding to H2O2 and phytohormone (Fig. 7). In summary, through analyses of genome-wide survey and expression profiling of GhAPX gene family, we provided some new insights in controlling H2O2 homeostasis during fiber development, that is decided by the ten fiber-preferentially accumulated GhAPXs.
In conclusion, we performed thoroughly investigation of upland cotton GhAPX gene family. The evolutionary analyses suggested a significant increase in evolutionary rate between the A-sub-genome duplicated paralogue genes GhAPX10A/11A/12A, while non-significant evolutionary rate between GhAPX10D/11D/12D. Tissue- and development-specific expression profiling of GhAPX genes revealed that 10 members were expressed in cotton fiber and GhAPX10A, GhAPX10D, GhAPX12A, and GhAPX12D showed high expression levels in 30-day fiber, while GhAPX1A/D, GhAPX3A/D, and GhAPX6A/D showed relative low expression levels. Together with the APX enzyme activity and H2O2 content assay, we demonstrated that different GhAPX family members are responsible for redox homeostasis during different cotton fiber development stages.
References
Alscher RG, Donahue JL, Cramer CL (1997) Reactive oxygen species and antioxidants: relationships in green cells. Physiol Plant 100(2):224–233
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34(Web Server issue):W369–W373
Chen ZJ, Guan X (2011) Auxin boost for cotton. Nat Biotechnol 29(5):407–409
Chew O, Whelan J, Millar AH (2003) Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem 278(47):46869–46877
Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407(6802):321–326
Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J et al (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17(1):268–281
Foyer CH, Halliwell B (1976) The presence of glutathione and glutathione reductase in chloroplasts: a proposed role in ascorbic acid metabolism. Planta 133(1):21–25
Fryer MJ, Ball L, Oxborough K, Karpinski S, Mullineaux PM, Baker NR (2003) Control of expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of leaves. Plant J 33(4):691–705
Guo J, Wu J, Ji Q, Wang C, Luo L, Yuan Y et al (2008) Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J Genet Genom 35(2):105–118
Guo K, Du X, Tu L, Tang W, Wang P, Wang M et al (2016) Fibre elongation requires normal redox homeostasis modulated by cytosolic ascorbate peroxidase in cotton (Gossypium hirsutum). J Exp Bot 67(11):3289–3301
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297
Huang J, Pang C, Fan S, Song M, Yu J, Wei H et al (2015) Genome-wide analysis of the family 1 glycosyltransferases in cotton. Mol Genet Genom 290:1805–1818
Jin X, Li Q, Xiao G, Zhu YX (2013) Using genome-referenced expressed sequence tag assembly to analyze the origin and expression patterns of Gossypium hirsutum transcripts. J Integr Plant Biol 55(7):576–585
Jin X, Wang L, He L, Feng W, Wang X (2016) Two-dimensional gel electrophoresis-based analysis provides global insights into the cotton ovule and fiber proteomes. Sci China Life Sci 59(2):154–163
Jin X, Zhu L, Yao Q, Meng X, Ding G et al (2017) Expression profiling of mitongen-activated protein kinase genes reveals their evolutionary and functional diversity in different rubber tree (Hevea brasiliensis) cultivars. Genes 8(10):E261
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D et al (2009) Circos: an information aesthetic for comparative genomics. Genom Res 19:1639–1645
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Li HB, Qin YM, Pang Y, Song WQ, Mei WQ, Zhu YX (2007) A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytol 175(3):462–471
Li F, Fan G, Wang K, Sun F, Yuan Y, Song G et al (2014) Genome sequence of the cultivated cotton Gossypium arboretum. Nat Genet 46(6):567–572
Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ et al (2015) Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol 33(5):524–530
Li R, Xin S, Tao C, Jin X, Li H (2017) Cotton ascorbate oxidase promotes cell growth in cultured tobacco bright yellow-2 cells through generation of apoplast oxidation. Int J Mol Sci 18(7):E1346
Liszkay A, van der Zalm E, Schopfer P (2004) Production of reactive oxygen intermediates (O2·–H2O2, and ·OH) by maize roots and their role in wall loosening and elongation growth. Plant Physiol 136(2):3114–3123
Mei W, Qin Y, Song W, Li J, Zhu Y (2009) Cotton GhPOX1 encoding plant class III peroxidase may be responsible for the high level of reactive oxygen species production that is related to cotton fiber elongation. J Genet Genomics 36(3):141–150
Najami N, Janda T, Barriah W, Kayam G, Tal M, Guy M et al (2008) Ascorbate peroxidase gene family in tomato: its identification and characterization. Mol Genet Genom 279(2):171–182
Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49:249–279
Panchuk II, Zentgraf U, Volkov RA (2005) Expression of the Apx gene family during leaf senescence of Arabidopsis thaliana. Planta 222(5):926–932
Pandey S, Fartyal D, Agarwal A, Shukla T, James D et al (2017) Abiotic stress tolerance in plants: myriad roles of ascorbate peroxidase. Front Plant Sci 8:581
Pang C-Y, Wang H, Pang Y, Xu C, Jiao Y, Qin Y-M, Western TL, Yu S-X, Zhu Y-X (2010) Comparative proteomics indicates that biosynthesis of pectic precursors is important for cotton fiber and root hair elongation. Mol Cell Proteomics 9(9):2019–2033
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D et al (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492(7429):423–427
Pei ZM, Murata Y, Benning G, Thomine S, Klüsener B, Allen GJ et al (2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406(6797):731–734
Petrov VD, Van Breusegem F (2012) Hydrogen peroxide-a central hub for information flow in plant cells. AoB Plants 2012:pls014. https://doi.org/10.1093/aobpla/pls014
Pinheiro C, Chaves MM (2011) Photosynthesis and drought: can we make metabolic connections from available data? J Exp Bot 62(3):869–882
Qin YM, Hu CY, Zhu YX (2008) The ascorbate peroxidase regulated by H2O2 and ethylene is involved in cotton fiber cell elongation by modulating ROS homeostasis. Plant Signal Behav 3(3):194–196
Qu AL, Ding YF, Jiang Q, Zhu C (2013) Molecular mechanisms of the plant heat stress response. Biochem Biophys Res Commun 432(2):203–207
Sharp KH, Mewies M, Moody PC, Raven EL (2003) Crystal structure of the ascorbate peroxidase-ascorbate complex. Nat Struct Biol 10(4):303–307
Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L et al (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18(3):651–664
Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y et al (2002) Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot 53(372):1305–1319
Smirnoff N (2000) Ascorbic acid: metabolism and functions of a multi-facetted molecule. Curr Opin Plant Biol 3(3):229–235
Suzuki N, Koussevitzky S, Mittler R, Miller G (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35(2):259–270
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
Tao C, Jin X, Zhu L, Li H (2016) Two-dimensional gel electrophoresis-based proteomic analysis reveals N-terminal truncation of the Hsc70 protein in cotton fibers in vivo. Sci Rep 6:36961. https://doi.org/10.1038/srep36961
Teixeira FK, Menezes-Benavente L, Margis R, Margis-Pinheiro M (2004) Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: inferences from the rice genome. J Mol Evol 59(6):761–770
Teixeira FK, Menezes-Benavente L, Galvão VC, Margis R, Margis-Pinheiro M (2006) Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta 224(2):300–314
Triplett BA, Kim HJ, Hinchliffe D, Sze SH, Thaxton P, Stelly D et al (2007) Phytohormone regulation of cotton fiber development in vitro. In: Biotechnology and sustainable agriculture 2006 and beyond: proceedings of the, Iaptc&b congress, august, pp 153–156
Wang K, Wang Z, Li F, Ye W, Wang J, Song G et al (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44(10):1098–1103
Wang L, Zhu Y, Hu W, Zhang X, Cai C, Guo W (2015) Comparative transcriptomics reveals jasmonic acid-associated metabolism related to cotton fiber initiation. PLoS One 10(6):e0129854. https://doi.org/10.1371/journal.pone.0129854
Wu H, Tian Y, Wan Q, Fang L, Guan X, Chen J et al (2017) Genetics and evolution of MIXTA genes regulating cotton lint fiber development. New Phytol. https://doi.org/10.1111/nph.14844
Xiao GH, Wang K, Huang G, Zhu YX (2016) Genome-scale analysis of the cotton KCS gene family revealed a binary mode of action for gibberellin A regulated fiber growth. J Integr Plant Biol 58(6):577–589
Xin S, Tao C, Li H (2016) Cloning and functional analysis of the promoter of an ascorbate oxidase gene from Gossypium hirsutum. PLoS One 11(9):e0161695. https://doi.org/10.1371/journal.pone.0161695
Yadav CB, Bonthala VS, Muthamilarasan M, Pandey G, Khan Y, Prasad M (2015) Genome-wide development of transposable elements-based markers in foxtail millet and construction of an integrated database. DNA Res 22:79–90
Yang YW, Bian SM, Yao Y, Liu JY (2008) Comparative proteomic analysis provides new insights into the fiber elongating process in cotton. J Proteome Res 7(11):4623–4637
Yao D, Wei Q, Xu W, Syrenne RD, Yuan JS, Su Z (2012) Comparative genomic analysis of NAC transcriptional factors to dissect the regulatory mechanisms for cell wall biosynthesis. BMC Bioinform 13(Suppl 15):S10. https://doi.org/10.1186/1471-2105-13-S15-S10
Zhang M, Zheng X, Song S, Zeng Q, Hou L, Li D, Zhao J, Wei Y, Li X, Luo M, Xiao Y, Luo X, Zhang J, Xiang C, Pei Y (2011) Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol 29(5):453–458
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J et al (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33(5):531–537. https://doi.org/10.1038/nbt.3207
Zhang X, Xu X, Yu Y, Chen C, Wang J, Cai C et al (2016) Integration analysis of MKK and MAPK family members highlights potential MAPK signaling modules in cotton. Sci Rep 6:29781
Zheng M, Meng Y, Yang C, Zhou Z, Wang Y, Chen B (2014) Protein expression changes during cotton fiber elongation in response to drought stress and recovery. Proteomics 14(15):1776–1795
Zhu YX (2016) The post-genomics era of cotton. Sci China Life Sci 59:109
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (Grant number 31660408) and Distinguished Youth innovation foundation of Bingtuan (Grant number 2014CD003).
Author information
Authors and Affiliations
Contributions
CT, LZ, and QX performed the experiments. CT and XJ performed the data analysis. CT, XJ, XW, and HL wrote the manuscript. HL designed and managed the project.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Previously reported APX1 gene/protein expression patterns in
G. hirsutum. Expression patterns of G. hirsutum APX1 gene/protein in different cotton fiber development stages were deduced from five previous references using qRT-PCR (Li et al. 2007; Zheng et al. 2014), northern blot (Guo et al. 2016), microarray (Shi et al. 2006), and two-dimensional electrophoresis (Yang et al. 2006). The expression levels of GhAPX1 in 0-day ovule and 5-day fibers were set to 1, according to the authors (TIF 253 KB)
Multiple sequence alignment of GhAPXs
. Multiple sequence alignment was performed using the protein sequences of the 26 GhAPXs. Conserved domains I, II, and III were underlined. The label “H” marked the heme-binding amino acids, “D” the distal cation-binding sites, “A” ascorbate binding amino acids, “C” the amino acids involved in formation of the catalytic site, “P” the proximal cation biding sites (TIF 1830 KB)
Expression analysis of
GhAPX genes using transcriptome and EST data. (a) The EST frequencies for each GhAPX gene were determined by screening over 0.3 million EST sequences downloaded from GenBank EST database. Higher EST hits numbers were considered to represent higher expression levels. Note the significant frequency difference between orthologs GhAPX11A and GhAPX11D, which is consistent with the qRT-PCR data shown in Fig. 4. (b) Heat map profiling of 26 GhAPX genes using RNA-seq data downloaded from genome database of G. hirsutum TM-1. The reads per kilobases per millionreads (RPKM) values were used to estimate the expression level of each GhAPX in different cotton tissues (TIF 1275 KB)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Tao, C., Jin, X., Zhu, L. et al. Genome-wide investigation and expression profiling of APX gene family in Gossypium hirsutum provide new insights in redox homeostasis maintenance during different fiber development stages. Mol Genet Genomics 293, 685–697 (2018). https://doi.org/10.1007/s00438-017-1413-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-017-1413-2