Abstract
This cross-sectional study assessed the prevalence of bacteria isolated from Spanish children with suspected chronic lower respiratory tract infection (LRTI) for whom bronchoalveolar lavage (BAL) was indicated. BAL fluid (BALF) was collected from 191 children (aged ≥ 6 months to < 6 years, with persistent or recurrent respiratory symptoms, non-responders to usual treatment) and cultured. Nasopharyngeal swabs (NPSs) were also obtained and cultured to assess concordance of BALF and NPS findings in the same patient. Streptococcus pneumoniae, Haemophilus influenzae, or Moraxella catarrhalis were identified from BALF with a bacterial load indicative of infection (> 104 colony-forming units/mL) in 10.5, 8.9, and 6.3% of children, respectively. Clinical characteristics were similar among participants, regardless of positivity status for any of the bacteria. Approximately 26% of pneumococcal isolates were PCV13 serotypes, and 96% of H. influenzae isolates were non-typeable (NTHi). Concordance between BALF and NPS isolates was 51.0% for S. pneumoniae, 52.1% for H. influenzae, and 22.0% for M. catarrhalis.
Conclusion: S. pneumoniae, NTHi, and M. catarrhalis were the main bacteria detected in BALF and NPS. Children with suspected chronic LRTI may benefit from a vaccine protecting against NTHi.
What is Known: • Chronic lower respiratory tract infection (LRTI) in children can cause high morbidity and is a major use of healthcare resources worldwide. Despite this, their etiology or potential preventive measures are poorly assessed. • Bronchoalveolar lavage can be used to determine bacterial etiology of chronic LRTI. |
What is New: • We used conventional and molecular techniques to show that Haemophilus influenzae, Streptococcus pneumoniae, and Moraxella catarrhalis were present in the LRT of Spanish children with suspected chronic LRTI • Concordance between isolates from bronchoalveolar lavage fluid and nasopharyngeal swabs was low, suggesting that samples from the upper respiratory tract could not reliably predict the bacterial etiology of suspected chronic LRTI |
Similar content being viewed by others
Introduction
Chronic cough is associated with significant morbidity and use of healthcare resources during childhood [7, 20, 31]. A major cause of chronic cough in children is chronic bacterial lower respiratory tract infection (LRTI) and, in particular, protracted bacterial bronchitis [8, 29, 30]. Multiple infectious agents are responsible for chronic LRTI [9], but its etiology is difficult to establish, mainly because invasive techniques are required to collect specimens. When indicated, bronchoalveolar lavage (BAL) can be used for the collection of samples from the lower respiratory tract. BAL fluid (BALF) cultures have been shown to be a reliable method for determining the bacterial etiology of chronic LRTI, with non-typeable Haemophilus influenzae (NTHi), Streptococcus pneumoniae, and Moraxella catarrhalis being among the most commonly identified pathogens [17, 18, 30, 40, 51]. Several studies assessed a possible correlation between BALF and nasopharyngeal swab (NPS) specimens in terms of chronic LRTI etiology, with inconsistent results [26,27,28]. However, if evidence of a correlation between NP and BAL cultures were found, NPS could potentially be used to predict the etiology of LRTI in children with chronic respiratory symptoms.
Pediatric vaccination has significantly reduced the prevalence and carriage of these pathogens if high-coverage immunization was achieved [1, 2, 49]. H. influenzae type b (Hib) vaccine was introduced in the Spanish national immunization program (NIP) in 2000 [12], and vaccine coverage was ≥ 92.9% in the last decade [34]. Although routine administration of pneumococcal conjugate vaccines (PCVs) has been recommended since 2001 in Spain, not all regions originally included PCVs in their official vaccination schedule. Since 2016, PCV vaccination has been included in the infant NIP for administration at 2, 4, and 11–12 months of age [12].
In this cross-sectional study, we identified and characterized bacteria found in BALF and NPS specimens taken from Spanish children aged ≥ 6 months to < 6 years with suspected chronic LRTI and for whom a BAL procedure had been indicated. The serotype distribution of S. pneumoniae and H. influenzae isolates detected was evaluated to determine if it was covered by currently available vaccines. This study also explored correlations between findings in BALF and NPS, in order to assess whether NPS could reliably predict the etiology of chronic LRTI.
Methods
Study design and participants
This epidemiological, cross-sectional study was conducted between September 2013 and September 2015 in seven hospitals in Spain. Enrolled children were aged ≥ 6 months to < 6 years, had clinical suspicion of chronic LRTI, and were recommended to undergo a fiberoptic bronchoscopy with BAL. Suspected chronic LRTI was defined as persistent or recurrent respiratory signs/symptoms not responding to usual treatment (receipt of bronchodilators and inhaled corticosteroids for at least 3 months, repeated cycles of antibiotics up to 15 days in duration): wet cough lasting > 4 weeks without additional associated symptoms, recurrent (≥ 3 episodes per year) or persistent (lasting > 3 months) wheezing, persistent pathologic auscultation (lasting > 4 weeks), or persistent (> 1 month) or recurrent (≥ 3 occurrences per year) infiltrates/atelectasis observed on chest radiograph. Exclusion criteria from enrolment were as follows: child in care, exacerbation of persistent respiratory symptoms or receipt of antibiotic treatment within 2 weeks prior to enrolment, known cystic fibrosis, immunosuppression, or other severe immunodeficiencies.
BALF was collected while children were under sedation with topical anesthesia, according to routine procedure [13], choosing the most most-affected lobe (as identified radiologically or by fiberoptic bronchoscopy) or the right middle lobe as the standard sampling lobe. The volume of lavage solution was calculated according to international guidance [13] and three samples were obtained for each BAL. NPS was taken at the same time the BAL was performed. Data on demographics and clinical characteristics were collected from interviews with parents or legally acceptable representatives (LARs), children’s medical records, and routine physical examinations performed during the visit.
The study was approved by Independent Ethics Committees from each hospital, and a written informed consent was obtained from the children’s parents or LARs. The study is registered at Clinicaltrials.gov (NCT02838407).
Study objectives
The primary objective was to identify and characterize the bacterial etiology in BALF specimens taken from children with suspected chronic LRTI, visiting the hospital for a BAL procedure. As previously described [26, 27, 30, 51], a bacterial load of S. pneumoniae, H. influenzae, or M. catarrhalis > 104 colony-forming units (CFU)/mL was considered indicative of infection when only one organism was identified in BALF; this cut-off was > 105 CFU/mL if more than one organism was present.
Secondary objectives included assessing the presence of bacterial pathogens with any bacterial load as detected by culture from BALF and NPS; characterizing S. pneumoniae, H. influenzae, and M. catarrhalis isolates identified in the upper and lower respiratory tract and assessing possible correlations between them; and describing the study population in terms of demographics and clinical characteristics.
Microbiology
The second aliquot of each wash was used for culture. Samples of ≥ 2 mL of BALF were stored in skim-milk tryptone glucose glycerol transport/storage medium [22]. NPS specimens were stored in the same medium [44]. All specimens were maintained at ≤− 70 °C until tested.
Pure and 10−3 to 10−5 dilutions of BALF and NPS specimens were seeded and duplicated in three bacteriological media: blood agar-gentamicin (5 mg/L) [45], chocolate agar-bacitracin (5000 U/L) [35] (incubated at 37 °C in 5% CO2), and MacConkey agar (incubated at 37 °C without CO2). Plates were observed at 24 and 48 h, and colony counting was performed to calculate the CFU/mL. Standard bacteriological methods (including Gram staining, catalase, oxidase, X and V factor requirement, optochin, bile solubility or DNAse test, depending of the phenotypic, and Gram stain characteristics of the colonies) were used to identify H. influenzae, S. pneumoniae, M. catarrhalis, and any other presumptive bacterial pathogens.
In addition, bacterial identification was confirmed by qualitative real-time polymerase chain reaction (RT-PCR) and bacterial load was determined by quantitative RT-PCR using lgtC, lytA, and copB genes for H. influenzae, S. pneumoniae, and M. catarrhalis, respectively [3, 6, 24, 25].
A duplex RT-PCR using lgtC and P6 genes as targets was performed to confirm the identification of NTHi and to differentiate from H. haemolyticus. S. pneumoniae serotyping was performed by passive agglutination test (Pneumotest latex kit) and confirmed by Quellung reaction using specific factor antisera (Statens Serum Institute, Copenhagen) or by PCR [37]. H. influenzae types were determined using polyvalent, a-, b-, c-, d-, e-, and f-specific antisera.
Susceptibility testing of H. influenzae, S. pneumoniae, and M. catarrhalis against penicillin, erythromycin azithromycin, tetracycline, levofloxacin, trimethoprim/sulfamethoxazole, and amoxicillin/clavulanate was performed by broth microdilution following the procedures recommended by The Clinical and Laboratory Standards Institute (CLSI) [11]. A β-lactamase test was performed to all H. influenzae and M. catarrhalis isolates using the commercial Nitrocephin [36] and the acidimetric [48] tests. To detect H. influenzae β-lactamase-non-producing ampicillin-resistant isolates, ampicillin resistance was tested by broth microdilution for all β-lactamase negative isolates. The classification of isolates as susceptible, intermediately susceptible, and resistant was based on minimum inhibitory concentrations according to the CLSI [11].
Two isolates of H. influenzae, S. pneumoniae, or M. catarrhalis selected per specimen type per child were compared to determine whether they were clones of each other or different (unique) organisms. Isolates were defined to be unique based on the comparison of serotype and antimicrobial susceptibility for S. pneumoniae and serotypable H. influenzae; uniqueness was defined based on the antibiogram alone for NTHi and M. catarrhalis. If the two isolates per specimen type per child had different serotypes, or the same serotype but different susceptibility patterns, they were considered unique and both were retained in the analyses.
Statistical analyses
Analyses were conducted on all evaluable children with available BALF or NPS specimens. Proportions of S. pneumoniae, H. influenzae, M. catarrhalis, and other bacteria detected by culture in BALF specimens and in NPS were computed with associated exact 95% confidence intervals (CIs). Serotype/type and/or antibiotic susceptibility distributions (number and percentage) were determined for isolates of S. pneumoniae, H. influenzae, and M. catarrhalis detected in BALF cultures meeting the cut-off for infection, and the same analyses were performed post hoc for BALF cultures with any bacterial load and NPS specimens. Concordance between BALF and NPS specimens for H. influenzae, S. pneumoniae, and M. catarrhalis was assessed descriptively as the number and percentage of unique isolates with positive results in both specimens for each bacterium.
Analyses were descriptive and were performed using the Statistical Analysis Systems v9.2.
Results
Study participants
A total of 197 children were enrolled, of whom six were eliminated from the analyses (two children not meeting the age criteria for inclusion, and four with receipt of antibiotic treatment within 2 weeks prior to enrollment). The mean age of participants was 39.8 months, and 48.2% were girls. In total, 88.0% of children had received ≥ 1 dose of any PCV and 72.3% had received ≥ 3 doses of PCV13; 95.3% of children had received ≥ 3 vaccine doses against Hib; and 80.1% had received antibiotics in an interval of time of < 6 months and > 2 weeks prior to enrollment (Table 1).
Culture and bacterial load
BALF
S. pneumoniae, H. influenzae, and M. catarrhalis were detected in cultures with any bacterial load in 30.5, 51.1, and 49.5% of children, respectively, and ≥ 1 of the three bacteria was detected by microbiological cultures in 74.2% of BALF specimens. S. pneumoniae, H. influenzae, and M. catarrhalis met the cut-off for infection in BALF specimens from 10.5, 8.9, and 6.3% of children, respectively (Table 2). These percentages were in line with those obtained by using by quantitative RT-PCR as method of detection. Co-infections with two and three of the three bacteria were detected in seven and zero children, respectively. Other bacterial organisms detected by culture growth were Staphylococcus aureus (in one child), Pseudomonas aeruginosa (in five children), Stenotrophomonas maltophilia (in four children), and Pseudomona putida (in two children).
NPS
S. pneumoniae, H. influenzae, and M. catarrhalis were detected in cultures from 51.8, 46.6, and 56.0% of children, respectively (Table 2). Other bacterial pathogens identified were S. aureus (in six children) and S. maltophilia (in one child).
Clinical characteristics
Similar frequencies of cough, wheezing, pathologic auscultation, and infiltrates/atelectasis were observed in children with or without BALF cultures indicative of infection with S. pneumoniae, H. influenzae, or M. catarrhalis; a similar observation was made for fibro-bronchoscopy indications and respiratory conditions (Table 3). The clinical characteristics and medical records for the children in whom the other bacteria were detected were similar to that of the entire study population.
Blood sample results were available for 109 (57.1%) children, for whom the mean white blood cell count was 540.2 × 109/L; the mean C-reactive protein level, 25.5 mg/L; and the mean erythrocyte sedimentation rate, 25.3 mm/h. A chest X-ray was done for 166 (86.9%) children; an abnormal result (images of persistent/recurrent atelectasis-consolidation in the same or different lobules) was detected in 143 (74.9%) of them, and for 122 (85.3%), the abnormality was considered by the investigator as relevant to the decision to collect BALF.
Serotype distribution
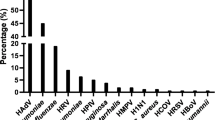
Among 65 unique isolates from BALF cultures with any bacterial load of S. pneumoniae, 10 (15.4%) were positive for serotype 18F, 17 (26.2%) for ≥ 1 PCV13 serotype (Fig. 1a, b and S1), and four (6.2%) for non-typeable S. pneumoniae. Among 21 S. pneumoniae isolates from BALF cultures meeting the cut-off for infection, 14 pneumococcal serotypes were identified, with serotype 18F having the highest prevalence (in four (19.0%) isolates). Six PCV13 vaccine serotypes (3, 6B, 14, 18C, 19A, and 19F) were present in 38.1% of these cultures (Fig. 1a).
Serogroup and serotype distribution of S. pneumoniae-positive children for cultures meeting the cut-off for infection from BALF (a; N = 21) and cultures of any load from NPS (b; N = 105); BALF, bronchoalveolar lavage fluid; NPS, nasopharyngeal swab; N, number of unique isolates from children with positive quantitative identification for S. pneumoniae; PHiD-CV, pneumococcal polysaccharide non-typeable Haemophilus influenzae protein D-conjugate vaccine; PCV13, 13-valent pneumococcal conjugate vaccines
Of 98 unique isolates from BALF cultures with any bacterial load of H. influenzae, 95 (96.9%) were positive for NTHi, and three (3.0%) for type f. Among the 18 unique H. influenzae isolates from cultures meeting the cut-off for infection, NTHi was identified in 17 (94.4%), while type f was detected in one (5.6%).
In NPS specimens, ≥ 1 PCV13 serotype was detected in 25.7% of isolates, and serotype 18F had the highest prevalence (13.3%) (Fig. 1b), while among the 92 isolates of H. influenzae, 95.7% were positive for NTHi and 4.3% for type f.
Antimicrobial susceptibility
Antimicrobial susceptibility of isolates from BALF and NPS specimens is presented in Table 4.
Concordance between BAL and NP isolates
Concordance between isolates from BALF and NPS specimens from children with positive bacterial cultures was established for 25 (51.0%) of 49 pneumococcal isolates, 37 (52.1%) of 71 H. influenzae isolates, and 18 (22.0%) of 82 M. catarrhalis isolates. Concordance between specimens meeting the cut-off for infection was lower (Tables 5 and 6).
Discussion
In this study in children aged ≥ 6 months to < 6 years with suspected chronic LRTI, S. pneumoniae was detected in 10.5% of BALF cultures with bacterial loads indicative of infection, while a prevalence of 8.9% for H. influenzae and 6.3% for M. catarrhalis was estimated. The observed prevalence of BALF findings indicative of infection and co-infection was lower than that reported in the literature.
There is a lack of consensus on a cut-off to differentiate a lower airway infection from upper respiratory tract contamination when culturing BALFs. Whereas 105 CFU/mL is the usual cut-off for pediatric cystic fibrosis patients [4], several studies propose values of 103 (in adult patients) or 104 CFU/mL [51] for pediatric chronic LRTI. Similar definitions for a load indicative of infection to the one used in our study was previously used [26, 27, 30, 51]. In a study in 30 non-indigenous young children with persistent respiratory symptoms in Australia using the same cut-off for infection, S. pneumoniae was identified as the major pathogen in the BALF of 23% of participants, followed by M. catarrhalis in 17% and NTHi in 7% of participants [26]. In another study in 197 children from the USA with persistent wet cough using ≥ 104 CFU/mL as a threshold for infection, NTHi was isolated in BALF cultures in 49% of children, while S. pneumoniae and M. catarrhalis were identified in 20 and 17% of children [51], in agreement with prior findings using the ≥ 105 CFU/mL cut-off [30]. Similar observations were made in 104 Indigenous Australian children with bronchiectasis, of whom 31% had NTHi infections, compared to 16% for S. pneumoniae and 12% for M. catarrhalis [27]. Bacterial co-infections were observed in our study in < 4% of BALF specimens, while prevalences of up to 35.4% have been reported [30, 40]. Our study did not identify any correlation between clinical characteristics of patients and presence of bacterial cultures with a load > 104 CFU/mL, likely due to the small sample size. However, our results confirmed previous findings establishing S. pneumoniae, NTHi, and M. catarrhalis as the main organisms in the BALF of children with suspected chronic LRTI. Of note, most children had received antibiotic treatment within 6 months prior to sampling, and this could have impacted the BALF results, especially if the antibiotic was taken within the interval of 2–4 weeks prior to procedure. This could explain the lower rates of infection and co-infection observed in our study compared to previous studies.
Although most children in our study received ≥ 2 doses of PCV13, serotypes 6B, 14, 18C, 19A, and 19F (common to both PCV13 and the pneumococcal polysaccharide protein D-conjugate vaccine PHiD-CV) and serotype 3 (PCV13 serotype) were present in 38.9% of BALF cultures with bacterial load indicative of infection and 26.2% of BALF cultures with any bacterial load. In NPS specimens, PCV13 serotypes were detected in 25.7% of isolates (serotypes 3, 6A, 6B, 19A, and 19F). Currently, two PCVs are used in pediatric immunization programs worldwide: a 10-valent PCV with cross-reactive protection to serotype 19A (PHiD-CV; [47]) and a 13-valent PCV (PCV13; [39]). Although PCV13 is recommended in the Spanish NIP, PHiD-CV is also authorized for use in children < 5 years of age. PHiD-CV has been shown to have a similar overall impact on invasive pneumococcal disease as PCV13 [15, 46], and the protein D from NTHi included as carrier component in PHiD-CV may provide additional protection against this organism [10]. Recent studies have reported a prevalence of up to 57% of PCV13 serotypes causing invasive disease from 2010 to 2014, in regions of Spain where vaccine coverage was estimated at around 55% [16, 33], together with an emergence of serotype 12F [16]. The most prevalent pneumococcal serotype identified in our study in both BALF and NPS specimens from children with suspected chronic LRTI was 18F, which is not included in either PCV. Serotype 18F is not frequently reported as a cause of invasive pneumococcal or mucosal disease, even in the post-PCV era [33]. The high prevalence of this serotype in our study may be due to local epidemiology/outbreak of a certain 18F strain, and genetic analyses are warranted to confirm this hypothesis.
In our study, H. influenzae was identified with bacterial loads indicative of infection in 8.9% of BALF specimens. The epidemiology of H. influenzae has changed following the widespread use of Hib vaccination during infancy, and NTHi has become more prevalent in both carriage and disease [50]. The introduction of PHiD-CV in routine childhood immunization schedule could help to protect children against NTHi carriage (and subsequent aspiration into lower respiratory tracts), since previous studies have shown a reduction of NTHi nasopharyngeal colonization in vaccine recipients compared with control groups [14, 41]. Moreover, PHiD-CV vaccination was associated with an improved immune response to NTHi in children with chronic suppurative lung disease [38].
A lack of concordance in findings from BALF and NPS specimens was evident in our study; the concordance was even poorer for specimens meeting the cut-off for infection, although the threshold of > 104 CFU/mL might not be relevant for NP carriage. These results seem to support previous observations that NP sampling of the upper respiratory tract does not necessarily correlate well with bacterial pathogens causing lower airway infections [4, 26, 42] and suggesting that NPS cannot replace BALF testing for children with suspected chronic LRTI. It is also possible that a combined assessment of NP and oropharyngeal microbiota of the upper respiratory tract would be better predictors of lower respiratory tract bacterial flora [32], bearing in mind that false positives due to contamination are possible with this type of specimens.
Most bacterial isolates detected in this study were susceptible to the tested antibiotics. This result was not surprising, as the high resistance rates previously reported in Spain [23] have fallen, and at present, S. pneumoniae, H. influenzae, and M. catarrhalis have low levels of resistance, similar to other European countries [19, 21, 43]. This observation is likely due to judicious use of antibiotics with respect to dosage, duration of treatment, and improved targeting of prescriptions for the correct indication.
Our study has several limitations. Most children received antibiotics within the 6 months prior to enrolment, which could have resulted in the low prevalence of bacteria from BALF meeting the diagnostic cut-off for infection and also potentially resulted in a lack of concordance between the findings from BALFs and NPs (e.g., if antibiotic treatment differently affected the BALF and NP flora). The BALF was collected from a single lung lobe, and neither BAL cytology nor measurement of inflammatory mediators was performed for these samples. In addition, the samples were not screened for viral pathogens which might have caused clinical presentations similar to those which constituted inclusion criteria for enrolment in our study; therefore, bacteria could not be definitely established as the only cause of the suspected LRTI. Although PCR assays can increase the sensitivity of organism detection vs. conventional culture techniques, to date, there is no PCR method which can unequivocally differentiate NTHi and H. haemolyticus [5], and this could constitute another potential limitation of our study.
Conclusions
S. pneumoniae, NTHi, and M. catarrhalis were identified as the main bacterial organisms in both BALF and NPS specimens from children with suspected chronic LRTI. Clinical characteristics were similar among study participants, regardless of the presence of any of the three pathogens in bacterial loads indicative of infections in BALF. Despite most participants being vaccinated with at least three PCV13 doses, PCV13 serotypes were recovered in approximately 25% of S. pneumoniae-positive BALF and NPS specimens.
A lack of concordance in findings in BALF and NPS from the same patient was observed, indicating that NPS cannot be used to predict the bacterial etiology in children with suspected chronic LRTI.
Future vaccination policies should take into account the etiology of chronic LRTI in children, and our results suggest that vaccination against NTHi disease may contribute to the prevention of LRTI.
Abbreviations
- BAL:
-
Bronchoalveolar lavage
- BALF:
-
Bronchoalveolar lavage fluid
- CFU:
-
Colony-forming units
- CI:
-
Confidence interval
- CLSI:
-
The Clinical and Laboratory Standards Institute
- LAR:
-
Legally acceptable representative
- LRTI:
-
Lower respiratory tract infection
- NIP:
-
National immunization program
- NPS:
-
Nasopharyngeal swab
- NTHi:
-
Non-typeable Haemophilus influenzae
- PCV:
-
Pneumococcal conjugate vaccine
- RT-PCR:
-
Real-time polymerase chain reaction
References
(2012) Pneumococcal vaccines WHO position paper—2012. Wkly Epidemiol Rec 87(14):129–144. http://www.who.int/wer/2012/wer8714/en/
(2013) Haemophilus influenzae type b (Hib) vaccination position paper—July 2013. Wkly Epidemiol Rec 88(39):413–426. http://www.who.int/wer/2013/wer8839/en/
Abdeldaim GM, Stralin K, Korsgaard J, Blomberg J, Welinder-Olsson C, Herrmann B (2010) Multiplex quantitative PCR for detection of lower respiratory tract infection and meningitis caused by Streptococcus pneumoniae, Haemophilus influenzae and Neisseria meningitidis. BMC Microbiol 10(1):310. https://doi.org/10.1186/1471-2180-10-310
Armstrong DS, Grimwood K, Carlin JB, Carzino R, Olinsky A, Phelan PD (1996) Bronchoalveolar lavage or oropharyngeal cultures to identify lower respiratory pathogens in infants with cystic fibrosis. Pediatr Pulmonol 21(5):267–275. https://doi.org/10.1002/(sici)1099-0496(199605)21:5<267::aid-ppul1>3.0.co;2-k
Binks MJ, Temple B, Kirkham LA, Wiertsema SP, Dunne EM, Richmond PC, Marsh RL, Leach AJ, Smith-Vaughan HC (2012) Molecular surveillance of true nontypeable Haemophilus influenzae: an evaluation of PCR screening assays. PLoS One 7(3):e34083. https://doi.org/10.1371/journal.pone.0034083
Carvalho Mda G, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, Steigerwalt A, Whaley M, Facklam RR, Fields B et al (2007) Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 45(8):2460–2466. https://doi.org/10.1128/jcm.02498-06
Chang AB, Robertson CF, Van Asperen PP, Glasgow NJ, Mellis CM, Masters IB, Teoh L, Tjhung I, Morris PS, Petsky HL, Willis C, Landau LI (2012) A multicenter study on chronic cough in children: burden and etiologies based on a standardized management pathway. Chest 142(4):943–950. https://doi.org/10.1378/chest.11-2725
Chang AB, Robertson CF, van Asperen PP, Glasgow NJ, Masters IB, Teoh L, Mellis CM, Landau LI, Marchant JM, Morris PS (2013) A cough algorithm for chronic cough in children: a multicenter, randomized controlled study. Pediatrics 131(5):e1576–e1583. https://doi.org/10.1542/peds.2012-3318
Chang AB, Upham JW, Masters IB, Redding GR, Gibson PG, Marchant JM, Grimwood K (2016) Protracted bacterial bronchitis: the last decade and the road ahead. Pediatr Pulmonol 51(3):225–242. https://doi.org/10.1002/ppul.23351
Clarke C, Bakaletz LO, Ruiz-Guiñazú J, Boyrs D, Mrkvan T (2017) Impact of protein D-containing pneumococcal conjugate vaccines on non-typeable Haemophilus influenzae acute otitis media and carriage. Exp Rev Vaccines 16(17):1–14. https://doi.org/10.1080/14760584.2017.1333905
Clinical and Laboratory Standards Institute (CLSI). M100-S25 Performance Standards for Antimicrobial Susceptibility Testing. January 2015
Comite Asesor de Vacunas de la Asociacion Espanola de Pediatria. Manual de vacunas en linea de la AEP. Calendarios de vacunacion en Espana. http://vacunasaep.org/documentos/manual/cap-7. Accesssed 28 February 2017
de Blic J, Midulla F, Barbato A, Clement A, Dab I, Eber E, Green C, Grigg J, Kotecha S, Kurland G, Pohunek P, Ratjen F, Rossi G (2000) Bronchoalveolar lavage in children. ERS Task Force on bronchoalveolar lavage in children. European Respiratory Society. Eur Respir J 15(1):217–231
de Carvalho CX, Kipnis A, Thorn L, de Andrade JG, Pimenta F, Brandileone MC, Zanella RC, Flannery B, Sgambatti S, Andrade AL (2011) Carriage of Haemophilus influenzae among Brazilian children attending day care centers in the era of widespread Hib vaccination. Vaccine 29(7):1438–1442. https://doi.org/10.1016/j.vaccine.2010.12.045
Deceuninck G, De Serres G, Boulianne N, Lefebvre B, De Wals P (2015) Effectiveness of three pneumococcal conjugate vaccines to prevent invasive pneumococcal disease in Quebec, Canada. Vaccine 33(23):2684–2689. https://doi.org/10.1016/j.vaccine.2015.04.005
Del Amo E, Esteva C, Hernandez-Bou S, Galles C, Navarro M, Sauca G, Diaz A, Gassiot P, Marti C, Larrosa N et al (2016) Serotypes and clonal diversity of Streptococcus pneumoniae causing invasive disease in the era of PCV13 in Catalonia, Spain. PLoS One 11(3):e0151125. https://doi.org/10.1371/journal.pone.0151125
Donnelly D, Critchlow A, Everard ML (2007) Outcomes in children treated for persistent bacterial bronchitis. Thorax 62(1):80–84. https://doi.org/10.1136/thx.2006.058933
Douros K, Alexopoulou E, Nicopoulou A, Anthracopoulos MB, Fretzayas A, Yiallouros P, Nicolaidou P, Priftis KN (2011) Bronchoscopic and high-resolution CT scan findings in children with chronic wet cough. Chest 140(2):317–323. https://doi.org/10.1378/chest.10-3050
European Center for Disease Prevention and Control: Antimicrobial resistance interactive database (EARS-Net); 2015. http://www.ecdc.europa.eu/en/healthtopics/antimicrobial_resistance/database/Pages/database.aspx. Accessed 14 March 2017
Faniran AO, Peat JK, Woolcock AJ (1998) Persistent cough: is it asthma? Arch Dis Child 79(5):411–414. https://doi.org/10.1136/adc.79.5.411
Garcia-Cobos S, Arroyo M, Perez-Vazquez M, Aracil B, Lara N, Oteo J, Cercenado E, Campos J (2014) Isolates of beta-lactamase-negative ampicillin-resistant Haemophilus influenzae causing invasive infections in Spain remain susceptible to cefotaxime and imipenem. J Antimicrob Chemother 69(1):111–116. https://doi.org/10.1093/jac/dkt324
Gibson LF, Khoury JT (1986) Storage and survival of bacteria by ultra-freeze. Lett Appl Microbiol 3(6):127–129. https://doi.org/10.1111/j.1472-765X.1986.tb01565.x
Gomez J, Ruiz-Gomez J, Hernandez-Cardona JL, Nunez ML, Canteras M, Valdes M (1994) Antibiotic resistance patterns of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis: a prospective study in Murcia, Spain, 1983-1992. Chemotherapy 40(5):299–303. https://doi.org/10.1159/000239210
Greiner O, Day PJ, Bosshard PP, Imeri F, Altwegg M, Nadal D (2001) Quantitative detection of Streptococcus pneumoniae in nasopharyngeal secretions by real-time PCR. J Clin Microbiol 39(9):3129–3134. https://doi.org/10.1128/JCM.39.9.3129-3134.2001
Greiner O, Day PJ, Altwegg M, Nadal D (2003) Quantitative detection of Moraxella catarrhalis in nasopharyngeal secretions by real-time PCR. J Clin Microbiol 41(4):1386–1390. https://doi.org/10.1128/JCM.41.4.1386-1390.2003
Hare KM, Grimwood K, Leach AJ, Smith-Vaughan H, Torzillo PJ, Morris PS, Chang AB (2010) Respiratory bacterial pathogens in the nasopharynx and lower airways of Australian indigenous children with bronchiectasis. J Pediatr 157(6):1001–1005. https://doi.org/10.1016/j.jpeds.2010.06.002
Hare KM, Leach AJ, Morris PS, Smith-Vaughan H, Torzillo P, Bauert P, Cheng AC, McDonald MI, Brown N, Chang AB, Grimwood K (2012) Impact of recent antibiotics on nasopharyngeal carriage and lower airway infection in Indigenous Australian children with non-cystic fibrosis bronchiectasis. Int J Antimicrob Agents 40(4):365–369. https://doi.org/10.1016/j.ijantimicag.2012.05.018
Hare KM, Smith-Vaughan HC, Chang AB, Pizzutto S, Petsky HL, McCallum GB, Leach AJ (2017) Propensity of pneumococcal carriage serotypes to infect the lower airways of children with chronic endobronchial infections. Vaccine 35(5):747–756. https://doi.org/10.1016/j.vaccine.2016.12.059
Kantar A (2016) Update on pediatric cough. Lung 194(1):9–14. https://doi.org/10.1007/s00408-015-9815-6
Marchant JM, Masters IB, Taylor SM, Cox NC, Seymour GJ, Chang AB (2006) Evaluation and outcome of young children with chronic cough. Chest 129(5):1132–1141. https://doi.org/10.1378/chest.129.5.1132
Marchant JM, Newcombe PA, Juniper EF, Sheffield JK, Stathis SL, Chang AB (2008) What is the burden of chronic cough for families? Chest 134(2):303–309. https://doi.org/10.1378/chest.07-2236
Marsh RL, Kaestli M, Chang AB, Binks MJ, Pope CE, Hoffman LR, Smith-Vaughan HC (2016) The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome 4(1):37. https://doi.org/10.1186/s40168-016-0182-1
Mendez-Lage S, Losada-Castillo I, Agulla-Budino A (2015) Streptococcus pneumoniae: serotype distribution, antimicrobial susceptibility, risk factors and mortality in Galicia over a two year-period. Enferm Infecc Microbiol Clin 33(9):579–584. https://doi.org/10.1016/j.eimc.2015.01.010
Ministry of Health, Social Services and Equality, Spain. Vaccine coverage. Statistical data. Available at http://www.msc.es/en/profesionales/saludPublica/prevPromocion/vacunaciones/coberturas.htm [last accessed March 2017]
Nye KJ, Fallon D, Gee B, Howe S, Messer S, Turner T, Warren RE (2001) A comparison of the performance of bacitracin-incorporated chocolate blood agar with chocolate blood agar plus a bacitracin disk in the isolation of Haemophilus influenzae from sputum. J Med Microbiol 50(5):472–475. https://doi.org/10.1099/0022-1317-50-5-472
O'Callaghan CH, Morris A, Kirby SM, Shingler AH (1972) Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother 1(4):283–288. https://doi.org/10.1128/AAC.1.4.283
Pai R, Gertz RE, Beall B (2006) Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol 44(1):124–131. https://doi.org/10.1128/jcm.44.1.124-131.2006
Pizzutto SJ, Yerkovich ST, Upham JW, Hales BJ, Thomas WR, Chang AB (2015) Improving immunity to Haemophilus influenzae in children with chronic suppurative lung disease. Vaccine 33(2):321–326. https://doi.org/10.1016/j.vaccine.2014.11.024
Prevnar 13 summary of product characteristics. http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/001104/WC500057247.pdf. Accesssed 28 February 2017
Priftis KN, Litt D, Manglani S, Anthracopoulos MB, Thickett K, Tzanakaki G, Fenton P, Syrogiannopoulos GA, Vogiatzi A, Douros K, Slack M, Everard ML (2013) Bacterial bronchitis caused by Streptococcus pneumoniae and nontypable Haemophilus influenzae in children: the impact of vaccination. Chest 143(1):152–157. https://doi.org/10.1378/chest.12-0623
Prymula R, Kriz P, Kaliskova E, Pascal T, Poolman J, Schuerman L (2009) Effect of vaccination with pneumococcal capsular polysaccharides conjugated to Haemophilus influenzae-derived protein D on nasopharyngeal carriage of Streptococcus pneumoniae and H. influenzae in children under 2 years of age. Vaccine 28(1):71–78. https://doi.org/10.1016/j.vaccine.2009.09.113
Rosenfeld M, Emerson J, Accurso F, Armstrong D, Castile R, Grimwood K, Hiatt P, McCoy K, McNamara S, Ramsey B, Wagener J (1999) Diagnostic accuracy of oropharyngeal cultures in infants and young children with cystic fibrosis. Pediatr Pulmonol 28(5):321–328. https://doi.org/10.1002/(SICI)1099-0496(199911)28:5<321::AID-PPUL3>3.0.CO;2-V
Sahm DF, Brown NP, Thornsberry C, Jones ME (2008) Antimicrobial susceptibility profiles among common respiratory tract pathogens: a GLOBAL perspective. Postgrad Med 120(3 Suppl 1):16–24. https://doi.org/10.3810/pgm.2008.09.suppl52.280
Satzke C, Turner P, Virolainen-Julkunen A, Adrian PV, Antonio M, Hare KM, Henao-Restrepo AM, Leach AJ, Klugman KP, Porter BD, Sá-Leão R, Scott JA, Nohynek H, O'Brien KL, WHO Pneumococcal Carriage Working Group (2013) Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 32(1):165–179. https://doi.org/10.1016/j.vaccine.2013.08.062
Schmid RE, Washington JA 2nd, Anhalt JP (1978) Gentamicin-blood agar for isolation of Streptococcus pneumoniae from respiratory secretions. J Clin Microbiol 7(5):426–427
Su WJ, Lo HY, Chang CH, Chang LY, Chiu CH, Lee PI, Lu CY, Hsieh YC, Lai MS, Lin TY (2016) Effectiveness of pneumococcal conjugate vaccines of different valences against invasive pneumococcal disease among children in Taiwan: a nationwide study. Pediatr Infect Dis J 35(4):e124–e133. https://doi.org/10.1097/inf.0000000000001054
Synflorix summary of product characteristics. http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/000973/WC500054346.pdf. Accesssed 28 February 2017
Thornsberry C, Kirven LA (1974) Ampicillin resistance in Haemophilus influenzae as determined by a rapid test for beta-lactamase production. Antimicrob Agents Chemother 6(5):653–654. https://doi.org/10.1128/AAC.6.5.653
Tsaban G, Ben-Shimol S (2017) Indirect (herd) protection, following pneumococcal conjugated vaccines introduction: a systematic review of the literature. Vaccine 35(22):2882–2891. https://doi.org/10.1016/j.vaccine.2017.04.032
Van Eldere J, Slack MP, Ladhani S, Cripps AW (2014) Non-typeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect Dis 14(12):1281–1292. https://doi.org/10.1016/S1473-3099(14)70734-0
Zgherea D, Pagala S, Mendiratta M, Marcus MG, Shelov SP, Kazachkov M (2012) Bronchoscopic findings in children with chronic wet cough. Pediatrics 129(2):e364–e369. https://doi.org/10.1542/peds.2011-0805
Acknowledgments
The authors thank the children and their parents for their participation in the study. They also thank Nathalie El Omeiri and Iris Villarreal who supported the study in its conceptual stage. The authors acknowledge Claire Verbelen and Petronela M. Petrar (XPE Pharma & Science c/o GSK Vaccines) for drafting the manuscript and Bart van Heertum (XPE Pharma & Science c/o GSK Vaccines) for manuscript coordination.
EPI-Strep-064 study group: Silvia Castillo Corullón, Maribel Barrio Gómez de Agüero, Manuel Sánchez Solís, Antonio Moreno Galdó.
Funding
GlaxoSmithKline Biologicals SA was the funding source and was involved in all stages of the study conduct and analysis. GlaxoSmithKline Biologicals SA also took responsibility for all costs associated with the development and publishing of the present manuscript.
Author information
Authors and Affiliations
Consortia
Contributions
AEM, JGL, JRVA, OAC, OSB, MSB, PML, ATV, MVD, JR, PGC, and CAT designed the study. AEM, JGL, JRVA, OAC, OSB, MSB, PML, ATV, YF, JR, CAT, and PGC acquired the data. AEM, JGL, MVD, JR, PGC, and CAT analyzed the data. AEM, JGL, OAC, OSB, MSB, PML, and ATV contributed to the conduct of the study. All authors participated in the interpretation of the data. All reviewed and revised the manuscript and approved the final manuscript as submitted.
Corresponding author
Ethics declarations
Ethical approval was granted by independent ethics committees from each study site.
Disclosure of potential conflicts of interest
AEM, JGL, JRVA, OAC, OSB, MSB, and ATV have no conflict of interest to declare. PML received grants from Vertex Pharmaceuticals and Gilead Sciences. YF, MVD, JR, PGC, and CAT are employees of the GSK group of companies and PGC owns restricted shares/stock options of the GSK group of companies.
Research involving human participants and/or animals
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Communicated by Nicole Ritz
Electronic supplementary material
ESM 1
BALF, bronchoalveolar lavage fluid; N, number of unique isolates from children with positive identification for S. pneumoniae (any load); PHiD-CV, pneumococcal polysaccharide non-typeable Haemophilus influenzae protein Dconjugate vaccine; PCV13, 13-valent pneumococcal conjugate vaccines (JPEG 89.6 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Escribano Montaner, A., García de Lomas, J., Villa Asensi, J.R. et al. Bacteria from bronchoalveolar lavage fluid from children with suspected chronic lower respiratory tract infection: results from a multi-center, cross-sectional study in Spain. Eur J Pediatr 177, 181–192 (2018). https://doi.org/10.1007/s00431-017-3044-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00431-017-3044-3