Abstract
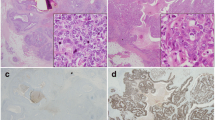
The TP63 gene encodes two major protein variants that differ in their N-terminal sequences and have opposing effects. In breast, ΔNp63 is expressed by immature stem/progenitor cells and mature myoepithelial/basal cells and is a characteristic feature of basal-like triple-negative breast cancers (TNBCs). The expression and potential role of TAp63 in the mammary gland and breast cancers is less clear, partly due to the lack of studies that employ p63 isoform-specific antibodies. We used immunohistochemistry with ΔNp63-specific or TAp63-specific monoclonal antibodies to investigate p63 isoforms in 236 TNBCs. TAp63, but not ΔNp63, was seen in tumour-associated lymphocytes and other stromal cells. Tumour cells showed nuclear staining for ΔNp63 in 17% of TNBCs compared to 7.3% that were positive for TAp63. Whilst most TAp63+ tumours also contained ΔNp63+ cells, the levels of the two isoforms were independent of each other. ΔNp63 associated with metaplastic and medullary cancers, and with a basal phenotype, whereas TAp63 associated with androgen receptor, BRCA1/2 wild-type status and PTEN positivity. Despite the proposed effects of p63 on proliferation, Ki67 did not correlate with either p63 isoform, nor did they associate with p53 mutation status. ΔNp63 showed no association with patient outcomes, whereas TAp63+ patients showed fewer recurrences and improved overall survival. These findings indicate that both major p63 protein isoforms are expressed in TNBCs with different tumour characteristics, indicating distinct functional activities of p63 variants in breast cancer. Analysis of individual p63 isoforms provides additional information into TNBC biology, with TAp63 expression indicating improved prognosis.



Similar content being viewed by others
References
Crum CP, McKeon FD (2010) p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu Rev Pathol 5:349–371
Nekulova M, Holcakova J, Coates P, Vojtesek B (2011) The role of p63 in cancer, stem cells and cancer stem cells. Cell Mol Biol Lett 16:296–327
Su X, Chakravarti D, Flores ER (2013) p63 steps into the limelight: crucial roles in the suppression of tumorigenesis and metastasis. Nat Rev Cancer 13:136–143
Rinne T, Spadoni E, Kjaer KW, Danesino C, Larizza D, Kock M, Huoponen K, Savontaus ML, Aaltonen M, Duijf P, Brunner HG, Penttinen M, van Bokhoven H (2006) Delineation of the ADULT syndrome phenotype due to arginine 298 mutations of the p63 gene. Eur J Hum Genet 14:904–910
van Bokhoven H, Hamel BC, Bamshad M et al (2001) p63 gene mutations in eec syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am J Hum Genet 69:481–492
Lehmann BD, Pietenpol JA (2014) Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol 232:142–150
Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW (2007) The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest 117:1370–1380
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10:515–527
Ribeiro-Silva A, Ramalho LN, Garcia SB, Brandao DF, Chahud F, Zucoloto S (2005) p63 correlates with both BRCA1 and cytokeratin 5 in invasive breast carcinomas: further evidence for the pathogenesis of the basal phenotype of breast cancer. Histopathology 47:458–466
Balboni AL, Hutchinson JA, DeCastro AJ et al (2013) DeltaNp63alpha-mediated activation of bone morphogenetic protein signaling governs stem cell activity and plasticity in normal and malignant mammary epithelial cells. Cancer Res 73:1020–1030
Giacobbe A, Compagnone M, Bongiorno-Borbone L, Antonov A, Markert EK, Zhou JH, Annicchiarico-Petruzzelli M, Melino G, Peschiaroli A (2016) p63 controls cell migration and invasion by transcriptional regulation of MTSS1. Oncogene 35:1602–1608
Holcakova J, Nekulova M, Orzol P, Nenutil R, Podhorec J, Svoboda M, Dvorakova P, Pjechova M, Hernychova L, Vojtesek B, Coates PJ (2017) DeltaNp63 activates EGFR signaling to induce loss of adhesion in triple-negative basal-like breast cancer cells. Breast Cancer Res Treat 163:475–484
Nekulova M, Holcakova J, Gu X, Hrabal V, Galtsidis S, Orzol P, Liu Y, Logotheti S, Zoumpourlis V, Nylander K, Coates PJ, Vojtesek B (2016) DeltaNp63alpha expression induces loss of cell adhesion in triple-negative breast cancer cells. BMC Cancer 16:782
Orzol P, Nekulova M, Holcakova J, Muller P, Votesek B, Coates PJ (2016) DeltaNp63 regulates cell proliferation, differentiation, adhesion, and migration in the BL2 subtype of basal-like breast cancer. Tumour Biol 37:10133–10140
Li N, Singh S, Cherukuri P, Li H, Yuan Z, Ellisen LW, Wang B, Robbins D, DiRenzo J (2008) Reciprocal intraepithelial interactions between TP63 and hedgehog signaling regulate quiescence and activation of progenitor elaboration by mammary stem cells. Stem Cells 26:1253–1264
Su X, Napoli M, Abbas HA, Venkatanarayan A, Bui NHB, Coarfa C, Gi YJ, Kittrell F, Gunaratne PH, Medina D, Rosen JM, Behbod F, Flores ER (2017) TAp63 suppresses mammary tumorigenesis through regulation of the hippo pathway. Oncogene 36:2377–2393
Svoboda M, Navratil J, Fabian P et al (2012) Triple-negative breast cancer: analysis of patients diagnosed and/or treated at the Masaryk memorial cancer institute between 2004 and 2009. Klin Onkol 25:188–198
Nenutil R, Smardova J, Pavlova S, Hanzelkova Z, Muller P, Fabian P, Hrstka R, Janotova P, Radina M, Lane DP, Coates PJ, Vojtesek B (2005) Discriminating functional and non-functional p53 in human tumours by p53 and MDM2 immunohistochemistry. J Pathol 207:251–259
Lucijanic M (2016) Survival analysis in clinical practice: analyze your own data using an Excel workbook. Croat Med J 57:77–79
Nekulova M, Holcakova J, Nenutil R, Stratmann R, Bouchalova P, Müller P, Mouková L, Coates PJ, Vojtesek B (2013) Characterization of specific p63 and p63-N-terminal isoform antibodies and their application for immunohistochemistry. Virchows Arch 463:415–425
Yoh KE, Regunath K, Guzman A, Lee SM, Pfister NT, Akanni O, Kaufman LJ, Prives C, Prywes R (2016) Repression of p63 and induction of EMT by mutant Ras in mammary epithelial cells. Proc Natl Acad Sci U S A 113:E6107–E6116
Orzol P, Holcakova J, Nekulova M, Nenutil R, Vojtesek B, Coates PJ (2015) The diverse oncogenic and tumour suppressor roles of p63 and p73 in cancer: a review by cancer site. Histol Histopathol 30:503–521
Bishop JA, Teruya-Feldstein J, Westra WH, Pelosi G, Travis WD, Rekhtman N (2012) p40 (DeltaNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod Pathol 25:405–415
Henderson SA, Torres-Cabala CA, Curry JL, Bassett RL, Ivan D, Prieto VG, Tetzlaff MT (2014) p40 is more specific than p63 for the distinction of atypical fibroxanthoma from other cutaneous spindle cell malignancies. Am J Surg Pathol 38:1102–1110
Lee JJ, Mochel MC, Piris A, Boussahmain C, Mahalingam M, Hoang MP (2014) p40 exhibits better specificity than p63 in distinguishing primary skin adnexal carcinomas from cutaneous metastases. Hum Pathol 45:1078–1083
Sailer V, Luders C, Kuhn W, Pelzer V, Kristiansen G (2015) Immunostaining of ΔNp63 (using the p40 antibody) is equal to that of p63 and CK5/6 in high-grade ductal carcinoma in situ of the breast. Virchows Arch 467:67–70
Wang X, Boddicker RL, Dasari S, Sidhu JS, Kadin ME, Macon WR, Ansell SM, Ketterling RP, Rech KL, Feldman AL (2017) Expression of p63 protein in anaplastic large cell lymphoma: implications for genetic subtyping. Hum Pathol 64:19–27
Aubry MC, Roden A, Murphy SJ, Vasmatzis G, Johnson SH, Harris FR, Halling G, Knudson RA, Ketterling RP, Feldman AL (2015) Chromosomal rearrangements and copy number abnormalities of TP63 correlate with p63 protein expression in lung adenocarcinoma. Mod Pathol 28:359–366
de Biase D, Morandi L, Degli Esposti R, Ligorio C, Pession A, Foschini MP, Eusebi V (2010) p63 short isoforms are found in invasive carcinomas only and not in benign breast conditions. Virchows Arch 456:395–401
Pruneri G, Fabris S, Dell'Orto P, Biasi MO, Valentini S, del Curto B, Laszlo D, Cattaneo L, Fasani R, Rossini L, Manzotti M, Bertolini F, Martinelli G, Neri A, Viale G (2005) The transactivating isoforms of p63 are overexpressed in high-grade follicular lymphomas independent of the occurrence of p63 gene amplification. J Pathol 206:337–345
Djelloul S, Tarunina M, Barnouin K, Mackay A, Jat PS (2002) Differential protein expression, DNA binding and interaction with SV40 large tumour antigen implicate the p63-family of proteins in replicative senescence. Oncogene 21:981–989
Provenzano E, Byrne DJ, Russell PA, Wright GM, Generali D, Fox SB (2016) Differential expression of immunohistochemical markers in primary lung and breast cancers enriched for triple-negative tumours. Histopathology 68:367–377
Thike AA, Cheok PY, Jara-Lazaro AR, Tan B, Tan P, Tan PH (2010) Triple-negative breast cancer: clinicopathological characteristics and relationship with basal-like breast cancer. Mod Pathol 23:123–133
Kurita T, Cunha GR, Robboy SJ, Mills AA, Medina RT (2005) Differential expression of p63 isoforms in female reproductive organs. Mech Dev 122:1043–1055
Nylander K, Vojtesek B, Nenutil R, Lindgren B, Roos G, Zhanxiang W, Sjöström B, Dahlqvist Å, Coates PJ (2002) Differential expression of p63 isoforms in normal tissues and neoplastic cells. J Pathol 198:417–427
Rampurwala M, Wisinski KB, O'Regan R (2016) Role of the androgen receptor in triple-negative breast cancer. Clin Adv Hematol Oncol 14:186–193
Yalcin-Ozuysal O, Fiche M, Guitierrez M, Wagner KU, Raffoul W, Brisken C (2010) Antagonistic roles of notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ 17:1600–1612
Buckley NE, Conlon SJ, Jirstrom K, Kay EW, Crawford NT, O'Grady A, Sheehan K, Mc Dade SS, Wang CW, McCance DJ, Johnston PG, Kennedy RD, Harkin DP, Mullan PB (2011) The {Delta}Np63 proteins are key allies of BRCA1 in the prevention of basal-like breast cancer. Cancer Res 71:1933–1944
Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C (2001) A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol 21:1874–1887
Muller PA, Vousden KH, Norman JC (2011) p53 and its mutants in tumor cell migration and invasion. J Cell Biol 192:209–218
Stindt MH, Muller PA, Ludwig RL, Kehrloesser S, Dotsch V, Vousden KH (2015) Functional interplay between MDM2, p63/p73 and mutant p53. Oncogene 34:4300–4310
Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW, Vogel H, Mills AA (2009) TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol 11:1451–1457
Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, el-Naggar A, Creighton CJ, Suraokar MB, Wistuba I, Flores ER (2010) TAp63 suppresses metastasis through coordinate regulation of dicer and miRNAs. Nature 467:986–990
Koster MI, Lu SL, White LD, Wang XJ, Roop DR (2006) Reactivation of developmentally expressed p63 isoforms predisposes to tumor development and progression. Cancer Res 66:3981–3986
Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, Biernaskie JA, Sinha S, Prives C, Pevny LH, Miller FD, Flores ER (2009) TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell 5:64–75
Su X, Gi YJ, Chakravarti D, Chan IL, Zhang A, Xia X, Tsai KY, Flores ER (2012) TAp63 is a master transcriptional regulator of lipid and glucose metabolism. Cell Metab 16:511–525
Iqbal J, Thike AA, Cheok PY, Tse GM, Tan PH (2012) Insulin growth factor receptor-1 expression and loss of PTEN protein predict early recurrence in triple-negative breast cancer. Histopathology 61:652–659
Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmström PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg Å, Parsons R (2005) PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res 65:2554–2559
Hu L, Liang S, Chen H, Lv T, Wu J, Chen D, Wu M, Sun S, Zhang H, You H, Ji H, Zhang Y, Bergholz J, Xiao ZXJ (2017) DeltaNp63alpha is a common inhibitory target in oncogenic PI3K/Ras/Her2-induced cell motility and tumor metastasis. Proc Natl Acad Sci U S A 114:E3964–E3973
Mitani Y, Li J, Weber RS, Lippman SL, Flores ER, Caulin C, El-Naggar AK (2011) Expression and regulation of the DeltaN and TAp63 isoforms in salivary gland tumorigenesis. Clin Exp findings Am J Pathol 179:391–399
Funding
This work was funded by grants MEYS-NPSI-LO1413 and Award LM15089 from the Ministry of Education, Youth and Sports, Czech Republic; GACR P206/12/G151 from the Grant Agency of the Czech Republic; and MH CZ-DRO (MMCI 00209805) from the Ministry of Health, Czech Republic. The funders did not have a role in the planning, performing or analysing the data, or in manuscript preparation or submission.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
All patient data were anonymised, and the study was performed retrospectively on redundant excess tissues following ethical approval by the Biobanking and Biomolecular resources Research Infrastructure (BBMRI) at MMCI, in accordance with European Union regulations and the Declaration of Helsinki.
Conflict of interest
BV is a consultant for and RN is a co-owner of Moravian Biotechnology, who produces the p63 isoform-specific antibodies used in this study. The company did not provide financial support or have any influence over the design or execution of the studies. All other authors declare that they have no conflicts of interest.
Rights and permissions
About this article
Cite this article
Coates, P.J., Nenutil, R., Holcakova, J. et al. p63 isoforms in triple-negative breast cancer: ΔNp63 associates with the basal phenotype whereas TAp63 associates with androgen receptor, lack of BRCA mutation, PTEN and improved survival. Virchows Arch 472, 351–359 (2018). https://doi.org/10.1007/s00428-018-2324-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-018-2324-2