Abstract
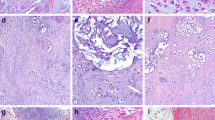
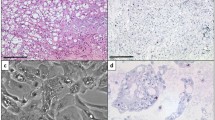
Chordomas are rare, slowly growing, locally aggressive bone neoplasms that arise from embryonic remnants of the notochord, showing dual epithelial-mesenchymal differentiation. The high plasticity probably is the main reason for the high variety in phenotypes of chordoma, from its high heterogeneity on a cellular level to its subtype variations depending on tissue location, with its potential to develop from an inactive quiescent form to an aggressive cancer with extreme adaptability and resistance to drugs and other treatments. Gene expression profiles of formalin-fixed, paraffin-embedded skull chordoma, spine chordoma, and normal tissue specimens were generated and compared. Using strict criteria, we identified 222 differentially expressed transcripts unique to skull base chordoma, 261 unique to spine chordoma, and 192 common to both chordoma subtypes. Further analysis of these three groups of transcripts allowed the selection of three subsets of highly differentially expressed genes as potential biomarkers, disease drivers, and therapeutic targets in both chordoma subtypes. Immunohistochemistry revealed LMX1A to be dominant in skull base chordoma, SALL3 to be unique to spine chordoma, and T to be common to both chordoma subtypes. In both chordoma subtypes, the genes with the highest expression were predominantly development-related genes, mostly transcription factors. Our findings indicate that these developmental genes play important oncogenic roles in chordoma, mainly causing high plasticity and resistance to therapy in both these cancer subtypes but also determining their differentiation status and proliferation activity, pointing to features expected of heterogeneous stem cell-like tissues with similarities to their notochord origins.




Similar content being viewed by others
References
Williams BJ, Raper DM, Godbout E et al (2013) Diagnosis and treatment of chordoma. J Natl Compr Cancer Netw 11:726–731
Chambers PW, Schwinn CP (1979) Chordoma. A clinicopathologic study of metastasis. Am J Clin Pathol 72:765–776
Higinbotham NL, Phillips RF, Farr HW, Hustu HO (1967) Chordoma. Thirty-five-year study at memorial hospital. Cancer 20:1841–1850
Bell D, Raza SM, Bell AH, Fuller GN, DeMonte F (2016) Whole-transcriptome analysis of chordoma of the skull base. Virchows Arch 469:439–449
Mardis ER (2014) Sequencing the AML genome, transcriptome, and epigenome. Semin Hematol 51:250–258
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Bell D, Bell AH, Bondaruk J, Hanna EY, Weber RS (2016) In-depth characterization of the salivary adenoid cystic carcinoma transcriptome with emphasis on dominant cell type. Cancer
Faial T, Bernardo AS, Mendjan S et al (2015) Brachyury and SMAD signalling collaboratively orchestrate distinct mesoderm and endoderm gene regulatory networks in differentiating human embryonic stem cells. Development 142:2121–2135
Chao TK, Yo YT, Liao YP et al (2013) LIM-homeobox transcription factor 1, alpha (LMX1A) inhibits tumourigenesis, epithelial-mesenchymal transition and stem-like properties of epithelial ovarian cancer. Gynecol Oncol 128:475–482
Yagi K, Satou Y, Satoh N (2004) A zinc finger transcription factor, ZicL, is a direct activator of Brachyury in the notochord specification of Ciona intestinalis. Development 131:1279–1288
Seifert A, Werheid DF, Knapp SM, Tobiasch E (2015) Role of Hox genes in stem cell differentiation. World J Stem Cells 7:583–595
Heng X, Breer H, Zhang X et al (2012) Sall3 correlates with the expression of TH in mouse olfactory bulb. J Mol Neurosci 46:293–302
Lin CC, Hsu YC, Li YH et al (2017) Higher HOPX expression is associated with distinct clinical and biological features and predicts poor prognosis in de novo acute myeloid leukemia. Haematologica 102:1044–1053
Li R, Campos J, Iida J (2015) A gene regulatory program in human breast cancer. Genetics 201:1341–1348
Iijima K, Yamada H, Miharu M et al (2012) ZNF385B is characteristically expressed in germinal center B cells and involved in B-cell apoptosis. Eur J Immunol 42:3405–3415
Sankar S, Yellajoshyula D, Zhang B, Teets B, Rockweiler N, Kroll KL (2016) Gene regulatory networks in neural cell fate acquisition from genome-wide chromatin association of Geminin and Zic1. Sci Rep 6:37412
Aruga J, Nozaki Y, Hatayama M, Odaka YS, Yokota N (2010) Expression of ZIC family genes in meningiomas and other brain tumors. BMC Cancer 10:79
Cha N, Liu W, Yang N et al (2014) Oncogenicity of LHX4 in colorectal cancer through Wnt/beta-catenin/TCF4 cascade. Tumour Biol 35:10319–10324
Bonnomet A, Polette M, Strumane K et al (2008) The E-cadherin-repressed hNanos1 gene induces tumor cell invasion by upregulating MT1-MMP expression. Oncogene 27:3692–3699
Draper BW, McCallum CM, Moens CB (2007) nanos1 is required to maintain oocyte production in adult zebrafish. Dev Biol 305:589–598
Rimkus TK, Carpenter RL, Qasem S, Chan M, Lo HW (2016) Targeting the sonic hedgehog signaling pathway: review of smoothened and GLI inhibitors. Cancers (Basel) 8
Corradetti MN, Inoki K, Guan KL (2005) The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem 280:9769–9772
Morquette B, Morquette P, Agostinone J et al (2015) REDD2-mediated inhibition of mTOR promotes dendrite retraction induced by axonal injury. Cell Death Differ 22:612–625
Yang YF, Pan YH, Cao Y et al (2017) PDZ binding kinase, regulated by FoxM1, enhances malignant phenotype via activation of beta-catenin signaling in hepatocellular carcinoma. Oncotarget
Li Q, Qi X, Jia W (2016) 3,3′,5-triiodothyroxine inhibits apoptosis and oxidative stress by the PKM2/PKM1 ratio during oxygen-glucose deprivation/reperfusion AC16 and HCM-a cells: T3 inhibits apoptosis and oxidative stress by PKM2/PKM1 ratio. Biochem Biophys Res Commun 475:51–56
Lorenzi L, Doring C, Rausch T et al (2017) Identification of novel follicular dendritic cell sarcoma markers, FDCSP and SRGN, by whole transcriptome sequencing. Oncotarget 8:16463–16472
Liu H, Zhang X, Wu J, French SW, He Z (2016) New insights on the palate, lung, and nasal epithelium clone (PLUNC) proteins: based on molecular and functional analysis of its homolog of YH1/SPLUNC1. Exp Mol Pathol 100:363–369
Ma Z, Li M, Roy S et al (2016) Chromatin boundary elements organize genomic architecture and developmental gene regulation in Drosophila Hox clusters. World J Biol Chem 7:223–230
Rhie SK, Guo Y, Tak YG et al (2016) Identification of activated enhancers and linked transcription factors in breast, prostate, and kidney tumors by tracing enhancer networks using epigenetic traits. Epigenetics Chromatin 9:50
Moorthy SD, Davidson S, Shchuka VM et al (2017) Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res 27:246–258
Mangum R, Varga E, Boue DR et al (2016) SHH desmoplastic/nodular medulloblastoma and Gorlin syndrome in the setting of Down syndrome: case report, molecular profiling, and review of the literature. Childs Nerv Syst 32:2439–2446
Funding source
This study was supported by start-up funds from MD Anderson (DB) and presented at AACR Advances in Sarcoma, Philadelphia May 2017.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Bell, A.H., DeMonte, F., Raza, S.M. et al. Transcriptome comparison identifies potential biomarkers of spine and skull base chordomas. Virchows Arch 472, 489–497 (2018). https://doi.org/10.1007/s00428-017-2224-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-017-2224-x