Abstract
The availability of a large number of whole-genome sequences allows comparative genomic analysis to reveal and understand evolution of regulatory regions and elements. The role played by events such as whole-genome and segmental duplications followed by genome fractionation in shaping genomic landscape and in expansion of gene families is crucial toward developing insights into evolutionary trends and consequences such as sequence and functional diversification. Members of Brassicaceae are known to have experienced several rounds of whole-genome duplication (WGD) that have been termed as paleopolyploidy, mesopolyploidy, and neopolyploidy. Such repeated events led to the creation and expansion of a large number of gene families. MIR319 is reported to be one of the most ancient and conserved plant MIRNA families and plays a role in growth and development including leaf development, seedling development, and embryo patterning. We have previously reported functional diversification of members of miR319 in Brassica oleracea affecting leaf architecture; however, the evolutionary history of the MIR319 gene family across Brassicaceae remains unknown and requires investigation. We therefore identified homologous and homeologous segments of ca. 100 kb, with or without MIR319, performed comparative synteny analysis and genome fractionation studies. We detected variable rates of gene retention across members of Brassicaceae when genomic blocks of MIR319a, MIR319b, and MIR319c were compared either between themselves or against Arabidopsis thaliana genome which was taken as the base genome. The highest levels of shared genes were found between A. thaliana and Capsella rubella in both MIR319b- and MIR319c-containing genomic segments, and with the closest species of A. thaliana, A. lyrata, only in MIR319a-containing segment. Synteny analysis across 12 genomes (with 30 sub-genomes) revealed MIR319c to be the most conserved MIRNA loci (present in 27 genomes/sub-genomes) followed by MIR319a (present in 23 genomes/sub-genomes); MIR319b was found to be frequently lost (present in 20 genomes/sub-genomes) and thus is under least selection pressure for retention. Genome fractionation revealed extensive and differential loss of MIRNA homeologous loci and flanking genes from various sub-genomes of Brassica species that is in accordance with their older history of polyploidy when compared to Camelina sativa, a recent neopolyploid, where the effect of genome fractionation was least. Finally, estimation of phylogenetic relationship using precursor sequences of MIR319 reveals MIR319a and MIR319b form sister clades, with MIR319c forming a separate clade. An intra-species synteny analysis between MIR319a-, MIR319b-, and MIR319c-containing genomic segments suggests segmental duplications at the base of Brassicaceae to be responsible for the origin of MIR319a and MIR319b.







Similar content being viewed by others
Reference
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408(6814):796–815
Arrigo N, Barker MS (2012) Rarely successful polyploids and their legacy in plant genomes. Curr Opin Plant Biol 15(2):140–146
Axtell MJ, Bowman JL (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13(7):343–349
Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16(7):1679–1691
Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13(2):137–144
Bowers JL, Chapman BA, Rong J, Paterson AH (2003) Unraveling angiosperms genome evolution by phylogenetic analysis of chromosomal duplications events. Nature 422(March):433–438
Bray N, Dubchak I, Pachter L (2003) AVID: a global alignment program. Genome Res 13(1):97–102
Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D (2003) The molecular analysis of leaf senescence—a genomics approach. Plant Biotechnol J 1(1):3–22
Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X et al (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345(6199):950–953
Cheng F, Wu J, Fang L, Sun S, Liu B, Lin K, Wang X (2012) Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One 7(5):e36442
Cheng F, Mandáková T, Wu J, Xie Q, Lysak MA, Wang X (2013) Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25(5):1541–1554
Cheng F, Wu J, Wang X (2014) Genome triplication drove the diversification of Brassica plants. Hortic Res 1:14024
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676
Conner JA, Conner P, Nasrallah ME, Nasrallah JB (1998) Comparative mapping of the Brassica S locus region and its homeolog in Arabidopsis: implications for the evolution of mating systems in the Brassicaceae. Plant Cell 10:801–881
Cusack BP, Wolfe KH (2007) When gene marriages don’t work out: divorce by subfunctionalization. Trends Genet 23(6):270–272
De Bodt S, Maere S, Van de Peer Y (2005) Genome duplication and the origin of angiosperms. Trends Ecol Evol 20(11):591–597
Edger PP, Pires JC (2009) Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosom Res 17(5):699–717
Fang L, Cheng F, Wu J, Wang X (2012) The impact of genome triplication on tandem gene evolution in Brassica rapa. Front Plant Sci 3:261
Fattash I, Voß B, Reski R, Hess WR, Frank W (2007) Evidence for the rapid expansion of microRNA-mediated regulation in early land plant evolution. BMC Plant Biol 7(1):13
Fei Y, Xue Y, Du P, Yang S, Deng X (2017) Expression analysis and promoter methylation under osmotic and salinity stress of TaGAPC1 in wheat (Triticum aestivum L). Protoplasma 254(2):987–996
Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151(4):1531–1545
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32:W273–W279
Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ et al (2013) An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet 45(8):891–898
Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM et al (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43(5):476–481
Jain A, Das S (2016) Synteny and comparative analysis of miRNA retention, conservation, and structure across Brassicaceae reveals lineage- and sub-genome-specific changes. Functional and Integrative Genomics 16(3):253–268
Jordon-Thaden I, Koch M (2008) Species richness and polyploid patterns in the genus draba (Brassicaceae): a first global perspective. Plant Ecol Divers 1(2):255–263
Kagale S, Robinson SJ, Nixon J, Xiao R, Huebert T, Condie J, Parkin IAP (2014a) Polyploid evolution of the Brassicaceae during the Cenozoic era. Plant Cell 26(7):2777–2791
Kagale S, Koh C, Nixon J, Bollina V, Clarke WE, Tuteja R et al (2014b) The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat Commun 5:3706
Kuittinen H, De Haan AA, Vogl C, Oikarinen S, Leppälä J, Koch M, Savolainen O (2004) Comparing the linkage maps of the close relatives Arabidopsis lyrata and Arabidopsis thaliana. Genetics 168(3):1575–1584
Lagercrantz U (1998) Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150(3):1217–1228
Langham RJ, Walsh J, Dunn M, Ko C, Goff SA, Freeling M (2004) Genomic duplication, fractionation and the origin of regulatory novelty. Genetics 166(2):935–945
Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14(5):836–843
Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA et al (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5:3930
Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinforma Oxf Engl 16:1046–1047
Mayrose I, Zhan SH, Rothfels CJ, Magnuson-Ford K, Barker MS, Rieseberg LH, Otto SP (2011) Recently formed polyploid plants diversify at lower rates. Science 333(6047):1257–1257
Murat F, Louis A, Maumus F, Armero A, Cooke R, Quesneville H, Salse J (2015) Understanding Brassicaceae evolution through ancestral genome reconstruction. Genome Biol 16(1):262
Nag A, King S, Jack T (2009) MIR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci U S A 106(52):22534–22539
Nagaharu U (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jap J Bot 7:389–452
Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Eshed Y (2007) Regulation of LANCEOLATE by MIR319 is required for compound-leaf development in tomato. Nat Genet 39(6):787–791
Osborn TC, Kole C, Parkin IAP, Sharpe AG, Kuiper M, Lydiate DJ, Trick M (1997) Comparison of flowering time genes in Brassica rapa, B napus and Arabidopsis thaliana. Genetics 146:1129–1129
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263
Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, Weigel D (2007) Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs MIR159 and MIR319. Dev Cell 13(1):115–125
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25:2383–2399
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Schnable JC, Wang X, Pires JC, Freeling M (2012) Escape from preferential retention following repeated whole genome duplications in plants. Front Plant Sci 3(May):1–8
Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Weigel D (2008) Control of jasmonate biosynthesis and senescence by MIR319 targets. PLoS Biol 6(9):e230
Schommer C, Bresso EG, Spinelli SV, Palatnik JF (2012) Role of microRNA MIR319 in plant development. In: Sunkar R (ed) MicroRNAs in plant development and stress responses. Springer, Berlin, Heidelberg, pp 29–47
Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11(11):535–542
Sharma A, Li X, Lim YP (2014) Comparative genomics of Brassicaceae crops. Breed Sci 64(1):3–13
Simillion C, Vandepoele K, Van Montagu MCE, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci 99(21):13627–13632
Singh NK, Anand S, Jain A, Das S (2017) Comparative genomics and synteny analysis of KCS17–KCS18 cluster across different genomes and sub-genomes of Brassicaceae for analysis of its evolutionary history. Plant Mol Biol Report 35(2):237–251
Singh S, Das S, Geeta R (2018) A segmental duplication in the common ancestor of Brassicaceae is responsible for the origin of the paralogs KCS6-KCS5, which are not shared with other angiosperms. Mol Phylogenet Evol 126:331–345
Slotte T, Hazzouri KM, Ågren JA, Koenig D, Maumus F, Guo YL et al (2013) The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet 45(7):831–835
Soltis PS, Liu X, Marchant DB, Visger CJ, Soltis DE (2014) Polyploidy and novelty: Gottlieb’s legacy. Phil Trans R Soc B 369(1648):20130351
Sun C, Wu J, Liang J, Schnable JC, Yang W, Cheng F, Wang X (2015) Impacts of whole genome triplication on MIRNA evolution in Brassica rapa. Genome Biol Evol 7(11):evv206
Sunkar R, Zhu JK (2004) Novel and stress-regulated MIRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2019–2186
Talmor-Neiman M, Stav R, Frank W, Voss B, Arazi T (2006) Novel micro-RNAs and intermediates of micro-RNA biogenesis from moss. Plant J 47(1):25–37
Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH (2008) Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res 18(12):1944–1954
Thiebaut F, Rojas CA, Almeida K, Grativol C, Domiciano GC, Lamb CRC, Ferreira PCG (2012) Regulation of MIR319 during cold stress in sugarcane. Plant Cell Environ 35(3):502–512
Thomas BC, Pedersen B, Freeling M (2006) Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res 16(7):934–946
Van der Graaff E (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141(2):776–792
Wang ZL, Li PH, Fredricksen M, Gong ZZ, Kim CS, Zhang C et al (2004) Expressed sequence tags from Thellungiella halophila, a new model to study plant salt-tolerance. Plant Sci 166(3):609–616
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Zhang Z (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43(10):1035–1039
Wang ST, Sun XL, Hoshino Y, Yu Y, Jia B, Sun ZW, Zhu YM (2014) MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS One 9(3):1–12
Warthmann N, Das S, Lanz C, Weigel D (2008) Comparative analysis of the MIR319a microRNA locus in Arabidopsis and related Brassicaceae. Mol Biol Evol 25(5):892–902
Warwick SI, Hall JC (2009) Phylogeny of Brassica and wild relatives (tribe Brassiceae: Brassicaceae) 3rd ed. Agric Agri-Food Res Branch Publ. Ottawa, ON, Canada. Contibution no. 991475.19–36
Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang X (2014) Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc Natl Acad Sci 111(14):5283–5288
Wu HJ, Zhang Z, Wang JY, Oh DH, Dassanayake M, Liu B et al (2012) Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc Natl Acad Sci 109(30):12219–12224
Yang R, Jarvis DJ, Chen H, Beilstein M, Grimwood J, Jenkins J et al (2013) The reference genome of the halophytic plant Eutrema salsugineum. Front Plant Sci 4:46
Yang J, Liu D, Wang X, Ji C, Cheng F, Liu B et al (2016) The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat Genet 48(10):1225–1232
Yu Y, Jia T, Chen X (2017) The ‘how’ and ‘where’ of plant microRNAs. New Phytol 216:1002–1017
Yunyan H, Bo L, Chao S, Jing L, Xiaobo W, Feng C, Jianli L, Xiaowu W, Jian W (2016) Evolution of TWIN SISTER of FT (TSF) genes in Brassicaceae. Horticultural Plant J 2(1):16–25
Zhang B, Pan X, Cobb GP, Anderson TA (2006) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289(1):3–16
Zhao W, Li Z, Fan J, Hu C, Yang R, Qi X, Wang S (2015) Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the MIR319/TCP4 module under root-knot nematode stress in tomato. J Exp Bot 66:4653–4667
Zhou M, Li D, Li Z, Hu Q, Yang C, Zhu L, Luo H (2013) Constitutive expression of a MIR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol 161(3):1375–1391
Zhu X, Leng X, Sun X, Mu Q, Wang B, Li X, Fang J (2015) Discovery of conservation and diversification of MIR171 genes by phylogenetic analysis based on global genomes. Plant Genome 8(2). https://doi.org/10.3835/plantgenome2014.10.0076
Acknowledgements
The research was supported by a Department of Biotechnology, Government of India (DBT) grant (no. BT/PR14532/AGR/36/673/2010) to S.D. G.J. would like to acknowledge JRF/SRF from University Grants Commission (UGC) for financial support; and C.C. to DBT for project Junior Research Fellowship (JRF). S.D. would also like to acknowledge financial assistance received from Delhi University under R&D grant support.
Author contribution statement
G.J. and S.D. were involved in planning of the study. G.J. collected all data and performed analysis along with C.C. G.J. and S.D. wrote the manuscript. All authors have read and approved of the manuscripts.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest statement
The authors declare that they have no conflict of interest.
Additional information
Communicated by Angelika Stollewerk
Electronic supplementary material
Supplementary Fig. 1
AVID/gVISTAs alignment of 100 kb region harboring MIR319a with A. thaliana whole genome. MIR319a is marked by red circle. (PPTX 13171 kb)
Supplementary Fig. 2
AVID/gVISTA alignment of 100 kb region harboring MIR319b with A. thaliana whole genome. MIR319b is marked by red circle. (PPTX 10233 kb)
Supplementary Fig. 3
AVID/gVISTA alignment of 100 kb region harboring MIR319c with A. thaliana whole genome. MIR319c is marked by red circle (PPTX 17208 kb)
Supplementary Fig. 4
Percentage of Arabidopsis thaliana homologs (y-axis) conserved across different genomic and sub-genomic segments (x-axis) harboring MIR319a, MIR319b and MIR319c in Brassicaceae. *Sub-genome not known (PNG 491 kb)
Supplementary Figure 5
Comparison of gene density as 1 gene / x kb (y-axis) in 100 kb region across different genomic and sub-genomic segments (x-axis) harboring MIR319a, MIR319b and MIR319c in Brassicaceae. *Sub-genome not known (PNG 513 kb)
Supplementary Figure 6
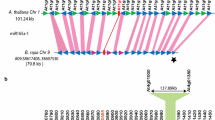
Effect of genome fractionation on two (B. rapa, B. oleracea, and B. napus) and three (C. sativa) sub-genomic homeologous segments that have retained or lost MIR319b. Genes present on all the three or any two sub-genomes are marked by black and blue arrows, respectively; green arrows marks genes unique to any sub-genomic segment. In B. rapa and B. napus, MIR319b (red arrow) is retained in LF and MF1 sub genome of all except B. oleracea and B. napus (C genome) and lost from all MF2 sub-genome; C. sativa retains MIR319b in all three sub genomic segments. (PNG 275 kb)
Supplementary Figure 7
Effect of genome fractionation on three (B. rapa, B. oleracea, and C. sativa) and six (B. napus) sub-genomic segments that have retained or lost MIR319c. Genes present on all the three or any two sub-genomes are marked by black and blue arrows, respectively; green arrows marks genes unique to any sub-genomic segment. In B. rapa, B. oleracea, and B. napus, MIR319c (red arrow) is retained in LF, MF1, and MF2 of all except B. oleracea and B. napus (C genome) LF sub-genome. C. sativa retains MIR319c in all sub genomic segments. (PNG 468 kb)
ESM 1
(DOCX 39 kb)
Supplementary Table 7
Detailed functional annotations of gene studied in microsynteny analysis of MIR319a, MIR319b and MIR319c (XLSX 72 kb)
Supplementary Table 8
Detailed functional annotations of gene studied in genome fractionation of MIR319a, MIR319b and MIR319c (XLSX 43 kb)
Rights and permissions
About this article
Cite this article
Joshi, G., Chauhan, C. & Das, S. Microsynteny analysis to understand evolution and impact of polyploidization on MIR319 family within Brassicaceae. Dev Genes Evol 228, 227–242 (2018). https://doi.org/10.1007/s00427-018-0620-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-018-0620-0