Abstract
Main conclusion
Comparative analysis of genome-wide miRNAs and their gene targets between cytoplasmic male sterile (CMS) and fertile lines of pigeonpea suggests a possible role of miRNA-regulated pathways in reproductive development.
Abstract
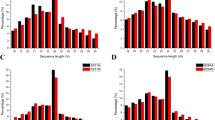
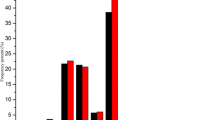
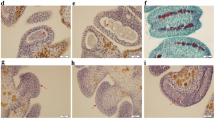
Exploitation of hybrid vigor using CMS technology has delivered nearly 50% yield gain in pigeonpea. Among various sterility-inducing cytoplasms (A1–A9) reported so far in pigeonpea, A2 and A4 are the two major sources that facilitate hybrid seed production. Recent evidence suggests involvement of micro RNA in vast array of biological processes including plant reproductive development. In pigeonpea, information about the miRNAs is insufficient. In view of this, we sequenced six small RNA libraries of CMS line UPAS 120A and isogenic fertile line UPAS 120B using Illumina technology. Results revealed 316 miRNAs including 248 known and 68 novel types. A total of 637 gene targets were predicted for known miRNAs, while 324 genes were associated with novel miRNAs. Degradome analysis revealed 77 gene targets of predicted miRNAs, which included a variety of transcription factors playing key roles in plant reproduction such as F-box family proteins, apetala 2, auxin response factors, ethylene-responsive factors, homeodomain-leucine zipper proteins etc. Differential expression of both known and novel miRNAs implied roles for both conserved as well as species-specific players. We also obtained several miRNA families such as miR156, miR159, miR167 that are known to influence crucial aspects of plant fertility. Gene ontology and pathway level analyses of the target genes showed their possible implications for crucial events during male reproductive development such as tapetal degeneration, pollen wall formation, retrograde signaling etc. To the best of our knowledge, present study is first to combine deep sequencing of small RNA and degradome for elucidating the role of miRNAs in flower and male reproductive development in pigeonpea.









Similar content being viewed by others
Abbreviations
- AGO:
-
Argonaute
- AP2:
-
Apetala 2
- ARF:
-
Auxin response factor
- Bp:
-
Base pair
- CDS:
-
Coding sequence
- CMS:
-
Cytoplasmic male sterility
- DCL:
-
Dicer-like
- GMUCT:
-
Genome-wide mapping of uncapped transcripts
- GO:
-
Gene ontology
- GRF:
-
Growth-regulating factor
- HD-Zip:
-
Homeodomain-leucine zipper
- miRNA:
-
Micro RNA
- MYB:
-
Myeloblastosis
- nc:
-
Non-coding
- NGS:
-
Next generation sequencing
- nt:
-
Nucleotide
- ORF:
-
Open reading frame
- PARE:
-
Parallel analysis of RNA ends
- PNRD:
-
Plant Non-coding RNA Database
- Pre-miRNA:
-
Precursor miRNA
- Pri-miRNA:
-
Primary miRNA
- Rf:
-
Fertility-restoring
- RISC:
-
RNA-induced silencing complex
- RLM-RACE:
-
RNA ligase-mediated rapid amplification of cDNA ends
- rRNA:
-
Ribosomal RNA
- SBP:
-
Squamosa promoter binding protein
- Sn RNA:
-
Small nuclear RNA
- Sno RNA:
-
Small nucleolar RNA
- sRNA:
-
Small RNA
- TCP:
-
Teosinte branched 1 cycloidea and PCF
- TF:
-
Transcription factor
- tRNA:
-
Transfer RNA
References
Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18:758–762
Addo-Quaye C, Miller W, Axtell MJ (2009) CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25:130–131
Alexa A, Rahnenführer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22:1600–1607
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Anthony AMFG (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17:705–721
Asha S, Sreekumar S, Soniya EV (2016) Unravelling the complexity of microRNA-mediated gene regulation in black pepper (Piper nigrum L.) using high-throughput small RNA profiling. Plant Cell Rep 35:53–63
Bai JF, Wang YK, Wang P et al (2017) Uncovering male fertility transition responsive mirna in a wheat photo-thermosensitive genic male sterile line by deep sequencing and degradome analysis. Front Plant Sci 8:1370
Bohra A, Mallikarjuna N, Saxena KB, Upadhyaya H, Vales I, Varshney R (2010) Harnessing the potential of crop wild relatives through genomics tools for pigeonpea improvement. J Plant Biol 37:83–98
Bohra A, Jha UC, Premkumar A, Bisht D, Singh NP (2016) Cytoplasmic male sterility (CMS) in hybrid breeding in field crops. Plant Cell Rep 35:967–993
Bohra A, Jha A, Singh IP, Pandey G, Pareek S, Basu PS, Chaturvedi SK, Singh NP (2017) Novel CMS lines in pigeonpea [Cajanus cajan (L.) Millspaugh] derived from cytoplasmic substitutions, their effective restoration and deployment in hybrid breeding. Crop J 5:89–94
Bohra A, Saxena KB, Varshney RK, Saxena RK (2020) Genomics assisted breeding for pigeonpea improvement. Theor Appl Genet 133:1721–1737
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 15:2114–2120
Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M (2008) Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20:1760–1774
Chen J, Zheng Y, Qin L, Wang Y, Chen L, He Y, Fei Z, Lu G (2016) Identification of miRNAs and their targets through high-throughput sequencing and degradome analysis in male and female Asparagus officinalis. BMC Plant Biol 16:80
Chen Z, Zhao N, Li S et al (2017) Plant mitochondrial genome evolution and cytoplasmic male sterility. Crit Rev Plant Sci 36:55–69
Dai X, Zhuang Z, Zhao PX (2018) psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 46(W1):W49–W54
Ding J, Zhou S, Guan J (2012) Finding MicroRNA targets in plants: current status and perspectives. Genom Proteom Bioinform 10:264–275
Ding X, Li J, Zhang H et al (2016) Identification of miRNAs and their targets by high throughput sequencing and degradome analysis in cytoplasmic male-sterile line NJCMS1A and its maintainer NJCMS1B of soybean. BMC Genom 17:24
Fang YN, Zheng BB, Wang L, Yang W, Wu XM, Xu Q, Guo WW (2016) High-throughput sequencing and degradome analysis reveal altered expression of miRNAs and their targets in a male-sterile cybrid pummelo (Citrus grandis). BMC Genomics 17:591
German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P et al (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26:941–946
Horn R, Gupta KJ, Colombo N (2014) Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 19:198–205
Jiang J, Lv M, Liang Y, Ma Z, Cao J (2014) Identification of novel and conserved miRNAs involved in pollen development in Brassica campestris ssp. chinensis by high-throughput sequencing and degradome analysis. BMC Genomics 15:146
Kompelli SK, Kompelli VSP, Enjala C, Suravajhala P (2015) Genome-wide identification of miRNAs in pigeonpea (Cajanus cajan L.). Aust J Crop Sci 9:215–222
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42(1):D68-73
Kravchik M, Stav R, Belausov E, Arazi T (2019) Functional characterization of microRNA171 family in tomato. Plants 8:10
Langmead B, Salzberg S (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Li X, Shahid MQ, Wu J, Wang L, Liu X, Lu Y (2016) Comparative small RNA analysis of pollen development in autotetraploid and diploid Rice. Int J Mol Sci 17:499
Li W, He Z, Zhang L, Lu Z, Xu J, Cui J, Wang L, Jin B (2017) miRNAs involved in the development and differentiation of fertile and sterile flowers in Viburnum macrocephalum f. keteleeri. BMC Genomics 18:783
Lin S, Su S, Jin L et al (2020) Identification of microRNAs and their targets in inflorescences of an Ogura-type cytoplasmic male-sterile line and its maintainer fertile line of turnip (Brassica rapa ssp. rapifera) via high-throughput sequencing and degradome analysis. PLoS ONE 15:0236829
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21:3448–3449
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12
Mishra A, Bohra A (2018) Non-coding RNAs and plant male sterility: current knowledge and future prospects. Plant Cell Rep 37:177–191
Nithin C, Thomas A, Basak J, Bahadur RP (2017) Genome-wide identification of miRNAs and lncRNAs in Cajanus cajan. BMC Genomics 18:1–14
Oesper L, Merico D, Isserlin R, Bader GD (2011) WordCloud: a Cytoscape plugin to create a visual semantic summary of networks. Source Code Biol Med 6:7
Omidvar V, Mohorianu I, Dalmay T, Fellner M (2015) Identification of miRNAs with potential roles in regulation of anther development and male-sterility in 7B–1 male-sterile tomato mutant. BMC Genomics 16:878
Reddy LJ, Faris DG (1981) A cytoplasmic-genetic male-sterile line in pigeonpea. Int Pigeonpea Newsl 1:16–17
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16:1616–1626
Ru P, Xu L, Ma H, Huang H (2006) Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res 16:457–465
Saxena KB, Sultana R, Mallikarjuna N, Saxena RK, Kumar RV, Sawargaonkar KL (2010) Male-sterility systems in pigeonpea and their role in enhancing yield. Plant Breed 129:125–134
Shen Y, Zhang Z, Lin H et al (2011) Cytoplasmic male sterility regulated novel microRNAs from maize. Funct Integr Genomics 11:179–191
Shi J, Dong M, Li L et al (2015) mirPRo - a novel standalone program for differential expression and variation analysis of miRNAs. Sci Rep 5:14617
Singh IP, Singh BB, Ali I, Kumar S (2009) Diversification and evaluation of cytoplasmic nuclear male sterility system in pigeonpea (Cajanus cajan). Indian J Agric Sci 79:291–294
Sinha P, Saxena KB, Saxena RK et al (2015) Association of nad7a gene with cytoplasmic male sterility in pigeonpea. Plant Genome 8:2
Sinha P, Singh VK, Saxena RK, Kale SM, Li Y, Garg V, Meifang T, Khan AW, Kim KD, Chitikineni A, Saxena KB, Sameer Kumar CV, Liu X, Xu X, Jackson S, Powell W, Nevo E, Searle IR, Lodha M, Varshney RK (2020) Genome-wide analysis of epigenetic and transcriptional changes associated with heterosis in pigeonpea. Plant Biotechnol J. https://doi.org/10.1111/pbi.13333
Thomson DW, Bracken CP, Goodall GJ (2011) Experimental strategies for microRNA target identification. Nucleic Acids Res 39:6845–6853
Tsuji H, Aya K, Ueguchi-Tanaka M et al (2006) GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J 47:427–444
Tuteja R, Saxena RK, Davila J, Shah T, Chen W, Xiao YL, Fan G, Saxena KB, Alverson AJ, Spillane C, Town C, Varshney RK (2013) Cytoplasmic male sterility-associated chimeric open reading frames identified by mitochondrial genome sequencing of four Cajanus genotypes. DNA Res 20:485–495
Ujino-Ihara T, Ueno S, Uchiyama K, Futamura N (2018) Comprehensive analysis of small RNAs expressed in developing male strobili of Cryptomeria japonica. PLoS ONE 13:e0193665
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687
Wei LQ, Yan LF, Wang T (2011) Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biol 12:R53
Wei M, Wei H, Wu M, Song M, Zhang J, Yu J, Fan S, Yu S (2013) Comparative expression profiling of miRNA during anther development in genetic male sterile and wild type cotton. BMC Plant Biol 13:66
Welchen E, Gonzalez DH (2005) Differential expression of the Arabidopsis cytochrome c genes Cytc-1 and Cytc-2 evidence for the involvement of TCP-domain protein binding elements in anther- and meristem-specific expression of the Cytc-1 gene. Plant Physiol 139:88–100
Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75:855–862
Yan J, Zhang H, Zheng Y, Ding Y (2015) Comparative expression profiling of miRNAs between the cytoplasmic male sterile line MeixiangA and its maintainer line MeixiangB during rice anther development. Planta 241:109–123
Yang J, Liu X, Xu B, Zhao N, Yang X, Zhang M (2013) Identification of miRNAs and their targets using high-throughput sequencing and degradome analysis in cytoplasmic male-sterile and its maintainer fertile lines of Brassica juncea. BMC Genom 14:9
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34(1):W293–W297
Yu JH, Zhao YX, Qin YT, Xiao HL (2013) Discovery of MicroRNAs associated with the S type cytoplasmic male sterility in maize. J Integr Agr 12:229–238
Yu D, Li L, Wei H, Yu S (2020) Identification and profiling of microRNAs and differentially expressed genes during anther development between a genetic male-sterile mutant and its wild type cotton via high-throughput RNA sequencing. Mol Genet Genomics 295:645–660
Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA (2006) Conservation and divergence of plant microRNA genes. Plant J 46:243–259
Zhang W, Xie Y, Xu L et al (2016a) Identification of microRNAs and their target genes explores miRNA mediated regulatory network of cytoplasmic male sterility occurrence during anther development in radish (Raphanus sativus L.). Front Plant Sci 7:1054
Zhang H, Hu J, Qian Q, Chen H, Jin J, Ding Y (2016b) Small RNA profiles of the rice PTGMS line Wuxiang S reveal miRNAs involved in fertility transition. Front Plant Sci 7:514
Zhang B, Zhang X, Liu G et al (2018) A combined small RNA and transcriptome sequencing analysis reveal regulatory roles of miRNAs during anther development of upland cotton carrying cytoplasmic male sterile Gossypium harknessii (D2) cytoplasm. BMC Plant Biol 18:242
Zhang H, Huang S, Tan J, Chen X, Zhang M (2020) MiRNAs profiling and degradome sequencing between the CMS-line N816S and its maintainer line Ning5m during anther development in pepper (Capsicum annuum L.). bioRxiv 1:1
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acid Res 31:3406–3415
Acknowledgements
AB acknowledges financial support from Indian Council of Agricultural Research (ICAR), New Delhi (Grant no.: AGENIASRICOP201501000047).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The author(s) declare that they have no competing interests.
Additional information
Communicated by Anastasios Melis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

425_2021_3568_MOESM7_ESM.png
Supplementary Fig 1 Gene ontology-Biological processes of genes (psRNATarget) targeted by known miRNAs. Color intensity represents the relative significance of the corresponding GO term (PNG 1184 KB)
Rights and permissions
About this article
Cite this article
Bohra, A., Gandham, P., Rathore, A. et al. Identification of microRNAs and their gene targets in cytoplasmic male sterile and fertile maintainer lines of pigeonpea. Planta 253, 59 (2021). https://doi.org/10.1007/s00425-021-03568-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-021-03568-6