Abstract
Main conclusion
Upregulation of the terpenoid pathway and increased ABA content in flax upon Fusarium infection leads to activation of the early plant’s response (PR genes, cell wall remodeling, and redox status).
Abstract
Plants have developed a number of defense strategies against the adverse effects of fungi such as Fusarium oxysporum. One such defense is the production of antioxidant secondary metabolites, which fall into two main groups: the phenylpropanoids and the terpenoids. While functions and biosynthesis of phenylpropanoids have been extensively studied, very little is known about the genes controlling the terpenoid synthesis pathway in flax. They can serve as antioxidants, but are also substrates for a plethora of different compounds, including those of regulatory functions, like ABA. ABA’s function during pathogen attack remains obscure and often depends on the specific plant-pathogen interactions. In our study we showed that in flax the non-mevalonate pathway is strongly activated in the early hours of pathogen infection and that there is a redirection of metabolites towards ABA synthesis. The elevated synthesis of ABA correlates with flax resistance to F. oxysporum, thus we suggest ABA to be a positive regulator of the plant’s early response to the infection.
Similar content being viewed by others
Introduction
Flax (Linum usitatissimum) is a valuable source of oil and fibers used in many industrial products. Its cultivation is restricted by environmental stress factors, but the biggest crop losses worldwide are caused by Fusarium infection, with Fusarium oxysporum f.sp. lini (Fol) being the most common. The Fusarium genus has a broad host range, although individual isolates usually only cause disease in a narrow range of plant species. The fungus is soil-borne and infection takes place mainly through the roots, although the hypocotyl may also be infected.
Fusarium oxysporum generally causes symptoms such as wilting, chlorosis, necrosis, premature leaf drop, and browning of the vascular system. Vascular wilt can account for up to 20% of the annual losses in flax cultivation. The fungus infects healthy plants by penetrating the root tips, root wounds, or lateral roots. It advances intracellularly into the xylem by producing microconidia, which germinate and thus block the vascular vessels, preventing water and nutrient translocation. This leads to epinasty and yellowing of the lower leaves, progressive wilting, and eventually death. After the plant dies, the fungus invades all of the tissues, sporulates, and infects neighboring plants (Kroes et al. 1998; Michielse and Rep 2009).
Most isolates of F. oxysporum do not produce mycotoxins, although there are reports that the fungi are able to synthesize moniliformin, beauvericin, diacetoxyscirpenol, T-2 toxin, HT-2 toxin, zearalenone, fumonisin and fusaric acid (Li et al. 2013). Mycotoxins can inhibit eukaryotic protein synthesis and facilitate the spread of the fungus because they inhibit plant defense mechanisms (Wagacha and Muthomi 2007). However, plants can produce compounds capable of interfering with mycotoxin production and they can degrade or detoxify trichothecenes by glycosylation (Karlovsky 2011; Shi et al. 2017).
The initial response of a plant to pathogen infection involves the production of reactive oxygen species (ROS). These can act directly against phytopathogen attack by killing the microorganism or limiting pathogen penetration of plant tissues by stiffening the cell wall. ROS achieve this by facilitating peroxidase reactions that catalyze intra- and inter-molecular cross-links between structural components of the cell wall and lead to lignin polymerization. In addition, ROS can act as signal molecules that activate plant cell defense mechanisms (De Gara et al. 2003). The reaction to the pathogen also involves production of pathogenesis-related (PR) proteins, including glucanases, chitinases and peroxidases, production of pectinase inhibitors, and changes in the cell wall, in particular due to pectin methylesterification and lignification, which create a physical barrier to fungal growth. ROS secretion must be strictly controlled due to the potentially harmful side effects, so plants also secrete antioxidative enzymes (superoxide dismutase, ascorbate peroxidase) and compounds (glutathione, secondary metabolites) (Wojtasik et al. 2011).
These secondary metabolites with antioxidative properties can be divided into two groups. Those produced via the phenylpropanoid pathway (phenolic acids, flavonoids, lignin and lignans) have been proven to participate in plant defense against pathogen invasion (Kostyn et al. 2012; Boba et al. 2016). The second group consists of isoprenoids, also called terpenoids. This large and most diverse class of compounds has a number of functions, including signaling and defense against pathogens.
The classification of terpenoids is based on the number of 5-carbon isoprene units, which constitute the molecule scaffold. Carotenoids, cytokinins, ABA, gibberellic acids, brassinosteroids, the phytol side chain of chlorophyll, and many more compounds, derive from terpenoid precursors. They serve a range of biological functions, acting as plant growth regulators, pigments, volatile attractants, and toxic deterrents in plant-animal interactions.
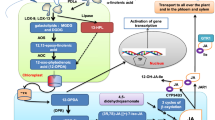
Two main pathways are employed in isoprene production: the mevalonate (MVA) pathway, which occurs in the cytosol of all types of organisms (Sonawane et al. 2016), and the non-mevalonate (MEP) pathway, which takes place in plastids (Lushchak and Semchuk 2012; Finkelstein 2013; Nisar et al. 2015) (Fig. 1). It is understood that the MVA pathway generates units for synthesis of 15- and 30-carbon terpenoids, such as squalene, sterols, stanols and brassinosteroids, while the MEP pathway leads to the synthesis of carotenoids (including xanthophylls), ABA, and gibberellins, and provides units for the synthesis of tocopherols and chlorophylls.
Scheme of the terpenoid biosynthesis pathway. DXP deoxy-d-xylulose 5-phosphate, MEP 2-C-methyl-d-erythritol 4-phosphate, IPP isopentenyl pyrophosphate, DMAPP dimethylallyl pyrophosphate, GPP geranyl pyrophosphate, FPP farnesyl pyrophosphate, GGPP geranylgeranyl pyrophosphate, DXS deoxy-d-xylulose 5-phosphate synthase, DXR deoxy-d-xylulose 5-phosphate reductase, IDI isopentenyl pyrophosphate isomerase, GPPS geranyl pyrophosphate synthase, PSY phytoene synthase, PDS phytoene desaturase, Z-ISO ζ-carotene isomerase, ZDS ζ-carotene desaturase, CRTISO carotenoid isomerase; LCYB lycopene-β cyclase, LCYE lycopene-ε cyclase, βHY β-carotene hydroxylase; ZEP zeaxanthin epoxidase, VDE violaxanthin de-epoxidase; NXS neoxanthin synthase, NCED 9-cis-epoxycarotenoid dioxygenase, ABA2 xanthoxin dehydrogenase, AAO abscisic aldehyde oxidase, CCD carotenoid cleavage dioxygenase, AACT acetyl CoA acetyltransferase, HMGS 3-hydroxy-3-methylglutaryl-CoA synthase, HMGR 3-hydroxy-3-methylglutaryl-CoA reductase, MVK mevalonate kinase, FPPS farnesyl pyrophosphate synthase, SQS squalene synthase, SQE squalene epoxidase, CAS cycloartenol cyclase, LAS lanosterol synthase, SMT1 sterol methyltransferase 1, SMO1 sterol-4alpha-methyl oxidase, COI cycloeucalenol cycloisomerase, CYP51G1 sterol 14-demethylase, FK Δ14 sterol reductase, HYD1 Δ8 isomerase, SMO2 sterol methyl oxidase, SMT2 sterol methyltransferase 2, STE1 Δ7-sterol-C5(6)-desaturase, CYP710a sterol C22 desaturase, GGR geranylgeranyl reductase; VTE2–HPT homogentisic acid phytyltransferase, VTE3 2-methyl-6-phytyl-1, 4-benzoquinol methyltransferase, VTE1 tocopherol cyclase, VTE4 γ-tocopherol methyltransferase. Enzymes of whose gene expression was not analyzed in this paper are indicated with a lighter color
Very little is known about the genes controlling the terpenoid synthesis pathway in flax. Only a few gene sequences had been presented in GenBank before genome sequencing projects were undertaken. Even now, a large amount of the data remains unannotated. On the other hand, the manipulation of terpenoid components in flax proved to be successful in increasing flax resistance to Fusarium infection (Boba et al. 2011). To understand the functioning of the terpenoid pathway in flax and provide the tools for genetic manipulation, we cloned the key genes of the flax terpenoid pathway and investigated their expression in response to Fusarium attack. Gene expression analysis results were correlated with the key compounds: carotenoids, tocopherols, sterols and ABA. The levels of callose and callose synthase gene expression, known to be regulated by ABA, and participating in plant defense responses, were assayed. To the best of our knowledge, this is the first report of a comprehensive study on the terpenoid pathway response in flax after F. oxysporum infection.
A particularly important role during a pathogen infection in plants is played by phytohormones and more specifically the interplay between their signaling. Salicylic acid (SA) is known to reduce plant susceptibility to pathogen infection, while jasmonic acid (JA), ethylene, ABA and auxin are involved in a more complex system of action, often exhibiting contradictory effects. All these hormones are part of a larger signaling network that integrates environmental inputs and provides a powerful system against microbial manipulations (Di et al. 2016). The inter-phytohormonal interactions are only partially understood, and especially the role of ABA in plant resistance to pathogens is controversial. According to current knowledge ABA influences some responses of plant pathogen resistance (stomatal closure, red-ox homeostasis), though its effect may vary with a number of variables, such as type of tissue, age of the plant, pathogen type, and stage of the infection (Maksimov 2009).
Under pathogenic F. oxysporum infection, the disease development is controlled by phytohormones, though the constant and correct mechanism of their actions cannot be defined, mainly due to the variable influence of different F. oxysporum strains. Thus, the ability of a particular strain to adapt to a particular host plant, which could explain the narrow host range observed in the F. oxysporum species, might be closely connected with the ability to manipulate the phytohormone signaling network. This hypothesis, although not proven yet, is strongly supported by some results on mutants with impaired hormone signaling, in which despite undisturbed colonization by the fungus, disease symptoms were significantly reduced (Thatcher et al. 2009; Cole et al. 2014). The current understanding is that the role of ABA in defense cannot be generalized as it appears to have a pathogen- and context-dependent role. Divergent ABA functions have been reported amongst the different necrotrophic species, including between B. cinerea, S. sclerotiorum, and A. brassicicola despite their common virulence and pathogenicity strategies. For example, in a study of tomato seedling–Alternaria solani interaction, ABA-treated plants showed improved resistance against infection (Song et al. 2011). Also, flax plants with increased ABA levels that were previously generated in our laboratory were more resistant to pathogen infection (Boba et al. 2018). However, the impaired ABA biosynthesis in Arabidopsis thaliana resulted in enhanced resistance to Plectosphaerella cucumerina (Sanchez-Vallet et al. 2012).
The first effect of ABA is closure of stomata to hinder further pathogen penetration. A group of ABA receptors (RCAR/PYR/PYL) was discovered recently which activate a cascade of responses after binding to the hormone (Cao et al. 2011; Lim and Lee 2015). Production of free radicals by NADPH oxidases and elevation of cellular calcium level are among the effects regulated by ABA (Mittler and Blumwald 2015). Transcriptome analysis of tomato after exogenous ABA application showed changes in the expression of many genes involved in the plant’s response to stress and infection. Among them, genes responsible for production and quenching of free radical and PR genes, including those connected with signal transduction through JA, were found (Wang et al. 2013). This is not surprising as about 15% of gene promoters possess ABA-responsive elements in A. thaliana (though they are not equally common in plant genomes of different species) (Gomez-Porras et al. 2007). A growing body of literature indicates that ABA is an indispensable element of the JA based response in some plant–pathogen interactions (Adie et al. 2007; García-Andrade et al. 2011).
Overall, ABA appears as a key regulator of defense against necrotrophs with both negative and positive contributions. Clarification of the nature of ABA function is further confounded by its interactions with other resistance pathways and the potential trade-offs resulting from the occurrence of abiotic stresses during infection (Ton et al. 2009).
Material and methods
Biological material and conditions of flax seedling growth
Flax seeds (Linum usitatissimum, var. Nike) were obtained from the Flax and Hemp Collection of the Institute of Natural Fibers in Poland. F. oxysporum f.sp. lini (MYA-1201) was obtained from the ATTC collection.
Flax seeds were sterilized in 50% plant preservative mixture PPM (Plant Cell Technology, Washington DC, USA) for 10 min and placed onto Petri dishes with MS medium supplemented with 0.9% agar. The dishes were then placed for 14 days in a phytotron, in a 16 h light (21 °C), 8 h darkness (16 °C) regime. 200 μl of F. oxysporum conidium suspension (1.76 × 107/ml) prepared as described by Di et al. (2017) was spread on the Petri dishes with potato dextrose agar (PDA) medium and cultivated for 2 days. The 14-day-old flax seedlings were transferred together with the MS medium onto the dishes with the fungi and after the appropriate time of induction (6, 12, 24, 36 or 48 h), the plants were collected, frozen in liquid nitrogen, and stored at − 80 °C for further analyses.
Reverse transcription real-time PCR analysis
Total RNA was isolated with Trizol according to the manufacturer’s protocol (Life Technologies). Genomic DNA was removed with DNase I as described in the manufacturer’s protocol (Thermo Scientific). RNA was transcribed to cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time RT-PCR was performed using the DyNAmo HS SYBR Green qPCR Kit (Thermo Scientific) on the Applied Biosystems StepOnePlus Real-Time PCR System. The primers used in the real-time RT-PCR are presented in Suppl. Table S1. The actin gene was used as a reference gene. The reaction conditions were: 95 °C for 15 min (holding stage); and 95 °C for 10 s, 57 °C for 20 s, 72 °C for 30 s, 37 cycles (cycling stage). The conditions of the melting curve stage were: 95 °C for 15 s, 60 °C for 1 min, 95 °C for 30 s, and ramp rate: 1.5%.
Isolation and analysis of terpenoids
The terpenoids were isolated according to the procedure described previously (Boba et al. 2018). Briefly, 25 mg of lyophilized seedling tissue was extracted sequentially with acetone, petroleum ether and diethyl ether for 10 min in an ultrasonic bath, and centrifuged at 10,000g and 4 °C for 10 min. The supernatants were collected, combined and dried under nitrogen and resuspended in 0.5 ml of methanol/acetonitrile mixture. The extraction was performed in three repetitions. The samples were then analyzed in UPLC with a 2996 PDA diode detector (Waters Acquity UPLC System) on a BEH C18 column, 2.1 × 100 mm, 1.7 μm (Waters Acquity). Eluent A was a mixture of 50% methanol, 40% acetonitrile and 10% H2O and eluent B was 50% methanol and 50% acetonitrile. The separation of the analyzed compounds was done at a 0.5 ml/min flow rate with the following program: 0–1 min 100% A, 2–9 min 100% B, 10 min 100% A. Identification of the components was done by comparison with the authentic standards (Sigma-Aldrich). The integration of peaks for carotenoids was done at 475 nm and for tocopherols at 290 nm.
Isolation and analysis of sterols
Three ml of methanol:chloroform (2:1, v/v) solution was added to 200 mg of plant tissue ground in liquid nitrogen and lyophilized and incubated at room temperature for 30 min. Then the samples were supplemented with 1 ml of chloroform and 1.8 ml of H2O, mixed and centrifuged. The chloroform layer was collected in a new tube and dried under nitrogen flow. The resulting pellet was placed in a glass tube with a PTFE cap and 2 ml of 2 M KOH and 30 µl of 5α-cholestane in hexane (internal standard) were added. Following thorough mixing the samples were incubated at 70 °C for 45 min. The samples were then cooled down and 1 ml of H2O, 0.5 ml of 96% ethanol and 1.5 ml of hexane were added. After shaking for 5 min (Multi Reax Shaker) the samples were centrifuged for 5 min at 500g. The hexane layer was collected in new tubes and the samples were extracted in the same manner twice more. The hexane fractions were combined, dried in nitrogen flow and silylated (BSTFA in 1% TMCS). The samples were shaken for 2 min and incubated at 70 °C for 45 min. The samples were dried under nitrogen flow again and re-suspended in 200 µl of hexane. Sterol analysis was performed with a gas chromatograph (Agilent Technologies 7890A) coupled with a flame ionization detector (FID). Qualitative and quantitative analysis was done based on comparing the obtained chromatograms with a library of chromatograms of pure standards.
Abscisic acid (ABA) isolation and quantification
Plant material was ground in a ceramic mortar at the temperature of liquid nitrogen directly before ABA isolation. 500 μl of extraction buffer (90% methanol, 200 μg/ml dithiocarb) was added to 200 mg of the ground plant material and mixed gently. The mixture was transferred to glass tubes and incubated at 4 °C overnight. Then the samples were centrifuged for 10 min at 6000g, 4 °C. The supernatant was transferred to a chilled Eppendorf tube and dried in a vacuum drier at 4 °C. The pellet was then resuspended in 400 μl of buffer containing 10% methanol, 50 mM Tris pH 8.0, 1 mM MgCl2, and 150 mM NaCl. The quantity of ABA was assayed with the Phytodetek Immunoassay Kit for ABA (Agdia Inc., Elkhart, IN, USA) according to the producer’s protocol.
Callose isolation and quantification
50 mg of lyophilized plant tissue was washed once with 96% ethanol and three times with 20% ethanol. Next 1 ml of 1 M NaOH was added to the tissue and incubated for 15 min at 80 °C. After subsequent centrifugation (15 min, 8000g) the supernatant was used for the callose assessment. 40 μl of 0.1% (w/v) aniline blue, 21 μl of 1 M HCl and 59 μl of 1 M glycine–NaOH (pH 9.5) buffer were added to 20 μl of the supernatant, mixed well and incubated for 20 min at 50 °C and for 30 min at room temperature. Callose content was measured spectrofluorometrically at 393 nm excitation wavelength and 484 nm emission wavelength. Curdlan (β-1,3-glucane) was used for standard curve preparation.
Dual staining of flax plants treated with ABA and infected with F. oxysporum
2-Week-old flax seedlings were subjected to 100 μM ABA treatment (sprayed all over the plants) and after 24 h growth they were infected with 1.3 × 105F. oxysporum conidia prepared as described by Di et al. (2017), and grown for another 4 h. Then the plants were collected and subjected to clearing with 0.15% TCA in ethanol:chloform mixture (4:1, v/v) for 48 h and then submitted to staining with safranin and solophenyl flavine 7GFE (0.1% w/v in 0.1 M Tris/HCl, pH 8.5) as follows. Whole plants were submerged in safranin solution (0.2% w/v safranin in 10% v/v ethanol) for 5 min and then washed three times in water. Subsequently, the plants were stained for 10 min with solophenyl flavine 7GFE (0.1% w/v in 0.1 M Tris/HCl, pH 8.5), and washed again with water (four times) (Knight and Sutherland 2011). Roots were cutoff and mounted on the slides, observed under the epi-fluorescent microscope Olympus BX50 using UV excitation light (360–370 nm) and documented using the Olympus DP71 camera and Cell^B software (Olympus Optical Co.). Images, taken at different depths, were processed using Helicon Focus 6.5.2 Pro (Helicon Soft Ltd.) and CorelDRAW 2017 (Corel Corporation).
Identification of fungal DNA in flax plants after the treatment with F. oxysporum
After the treatment with F. oxysporum, roots of the flax plant (50 mg) were used for DNA isolation with DNeasy Plant Mini Kit (Qiagen), which then served as the matrix in the RT-PCR. A 150 bp fragment of fungal murein transglycosylase gene was multiplied with the following primers: Fwd: TCTCAACGGTGTCGAGTCTAA and Rev: CACCCTGGTTGCAGATAAT. Actin gene was used for reference.
Treatment of F. oxysporum with ABA
To determine the impact of abscisic acid on F. oxysporum growth PDA media supplemented with ABA in five concentrations (0.1 μM, 0.5 μM, 1 μM, 10 μM and 50 μM) were prepared. Fungi (1.76 × 105 conidia) were grown on PDA for 48 h in darkness and compared to the control (PDA with appropriate amounts of solvent).
Statistical analysis
The experiments were performed in three repetitions. The statistical significance was calculated with Tukey’s test and with the Kruskal–Wallis test. The differences were statistically significant at P < 0.05.
Results
Identification of the partial cDNA sequences of genes involved in the carotenoid, tocopherol, and sterol biosynthesis pathways
To ascertain the roles of the main terpenoids found in flax carotenoids, sterols and tocopherols in response to F. oxysporum infection, we analyzed the expression levels of the genes of the above-mentioned pathways. Sequences of some of the genes were known. The other partial cDNA sequences were isolated via PCR with primers designed to the most homologous regions from other plants.
Analysis of transcript levels of genes involved in isoprenoid biosynthesis
Isoprenoids are synthesized in two main routes in plants, namely the cytosolic (mevalonate) and plastidial (non-mevalonate) routes. To verify which of the above-mentioned routes is activated upon Fusarium infection, we analyzed the transcript levels of the key genes of the routes, at 6, 12, 24, 36 and 48 h post infection (hpi).
We observed a general decrease of the transcript levels of the genes involved in the terpenoid backbone synthesis, both of the cytosolic and plastidial branches. Only for DXR and HMGR gene transcript levels an increase was detected from 12 hpi on (Fig. 2a). The decrease of the transcript levels of genes involved in the subsequent steps of the cytosolic part of the terpenoid biosynthesis pathway, i.e., leading to phytosterols, was even more pronounced (Fig. 2b).
Among the sterols identified in flax seedlings treated with the pathogens (squalene, campesterol, stigmasterol, β-sitosterol, sitostanol and avenasterol), the levels of the most abundant (campesterol and β-sitosterol) did not change at the time points investigated, while a consistent decrease in the levels of the remaining sterols was observed (Fig. S1).
A general increase in the transcript levels of genes involved in tocopherol synthesis was observed throughout the studied time points. Only at 36 hpi a clear decrease was noted for all of the genes investigated (Fig. 2c). The content of α-tocopherol did not change considerably throughout all time points analyzed. A small decrease was noted at 6 hpi (11% below the control) and a slight increase at subsequent time points, reaching 12% of the control at 48 hpi (Fig. S2).
As early as after 6 hpi, activation of the majority of the genes involved in the carotenoid synthesis occurred (Fig. 2d). This activation remained in most cases at all the time points investigated. A noticeable exception was the LCB gene, whose expression level decreased by ~ 25% in the early hours of infection, but returned to the control level at 48 hpi.
Neoxanthin, violaxanthin, lutein and β-carotene levels were assayed in flax seedlings treated with the pathogen. All the investigated compound levels decreased at 6 hpi (max. by 19%) to reach a moderate increase at 12 hpi (max. by 22% for violaxanthin) and a further decrease to the control level (except β-carotene, whose level dropped by 11% at 36 and 48 hpi), but no considerable changes were observed (Fig. S3).
Analysis of expression levels of genes of ABA biosynthesis route and abscisic acid content in response to F. oxysporum infection
Expression of seven genes involved in carotenoid degradation and ABA biosynthesis was determined in seedlings treated with the pathogen, namely CCD1, CCD7, CCD8, NCED3, NCED6, AAO and ABA2. A general activation of the genes connected with the ABA synthesis was observed, while the transcript levels of the CCD genes, leading to other apocarotenoids, decreased. The transcript level of the CCD1 was only highly elevated at 6 hpi, but then followed by a gradual decrease. A considerable increase in the transcript levels of the NCED3 and NCED6 genes was observed. Following the initial inertia at 6 hpi, about two-threefold elevation was noted at 12 hpi, 24 hpi and 36 hpi, to reach a sevenfold (NCED3) and tenfold (NCED7) increase at 48 hpi. AAO expression increased slowly to twofold of the control at 36 hpi (Fig. 3). Altogether, the data reveal activation of the ABA synthesis route after pathogen infection (Fig. 2e).
Transcript level of genes involved in apocarotenoid biosynthesis in flax after Fusarium oxysporum infection presented as RQ relatively to control (non-infected; RQ = 1). Actin gene was used for reference. Results are presented as mean values ± SD (n = 3). Statistically significant differences (P < 0.05) are indicated with asterisks
To examine whether the increased expression of genes involved in ABA synthesis corresponds with a higher content of this compound, we measured the level of this phytohormone in seedlings after pathogen infection. An elevated ABA level was observed starting from 12 hpi (threefold) and its maximum was achieved at 36 hpi (7.7-fold compared to the control) (Fig. 4).
Analysis of expression level of callose synthase gene and callose content in response to F. oxysporum infection
Abscisic acid regulates the expression of many genes, including callose synthase (PMR4), of which participation during the plant’s response to pathogen infection has been confirmed. At almost all time points (except at 24 hpi) an increase in gene expression was observed, yet the changes were only at 36 hpi statistically significant (Fig. 5a). Increased expression of PMR4 translated to an elevated level of callose (2.7-fold that of the control at 48 hpi) (Fig. 5b).
a Level of callose synthase (PMR4) gene transcript presented as RQ relative to control (non-treated with F. oxysporum; RQ = 1 − marked with black line). Actin gene was used for reference. Results are presented as mean values ± SD (n = 3). b Level of callose in flax seedlings infected with F. oxysporum (grey bars) presented as means of three independent repeats ± SD compared to the control (black bars). Statistically significant (P < 0.05) differences are indicated with asterisks
Staining of roots of flax treated with ABA after F. oxysporum infection
To determine the influence of ABA on fungal progression we performed microscopic observation of flax roots infected with F. oxysporum 24 h after treatment with ABA. The time was selected based on our previous studies where we observed high expression of the chitinase gene (PR gene) at this time point (Boba et al. 2018). We observed much lower penetration of flax root cells by fungal hyphae in the samples after ABA treatment compared to the control (Fig. 6).
a, b Visualization of Fusarium hyphae infesting flax roots 4 h after infection. Numerous hyphae form a network at the root surface. Hyphae, stained with solophenyl flavine 7GFE, show bright cyan fluorescence in UV exciting light. c, d Flax roots treated with 100 μM ABA 4 h after infection. Scale bars = 200 µm
Detection of fungal DNA in the flax roots after F. oxysporum treatment
Fungal DNA presence in the flax plant tissue after F. oxysporum treatment was confirmed after 2 h, 4 h, and 6 h of infection (Fig. 7). In the ABA-treated plants a not statistically significant increase of the fungal DNA level at 2 hpi was observed, followed by a decrease at 4 hpi and 6 hpi in comparison to the plants without ABA treatment.
Transcript level of fungal murein transglycosylase gene measured in flax tissue treated with 100 μM ABA collected after 2 h, 4 h, and 6 h of F. oxysporum infection in relation to the ABA non-treated control (RQ = 1, marked with line). Actin gene was used for reference. Results are presented as mean values ± SD (n = 3). Statistically significant differences (P < 0.05) are indicated with asterisks
Influence of ABA on F. oxysporum growth
To investigate the direct effect of ABA on fungal growth we treated F. oxysporum with different concentrations of abscisic acid. We did not observe any changes in the fungus growth or appearance. The areas of the fungal mycelia were comparable with the control (data not shown).
Discussion
There is a very limited amount of data on carotenoid pathway activation in response to infection. On the contrary, there are some reports where the carotenoid synthesis genes were downregulated, suggesting that isoprene units flux towards synthesis of tocopherols and monoterpenoid-derived phytoalexins (Gonzalez-Candelas et al. 2010). Hence, discovery of this pathway’s activation in flax was of great interest, as it seemed contradictory to all previously published results. In fact, there is only one report on carotenoid increase in sesame plants (Radhakrishnan et al. 2013). However, all of the examples concern different plants and different pathogens, suggesting that the response of plants seems quite plant-pathogen specific.
In our study we found that the levels of carotenoids in flax tissues did not change significantly in the early hours of F. oxysporum infection, which, however, did not correlate with the level of transcripts of the genes involved in their synthesis. Activation of the plastidial part of terpenoid biosynthesis occurs as early as at 6 hpi and is accompanied by decreased activity of the phytosterol synthesis genes. Phytosterols are known to increase plant resistance to pathogens, but it appears that this occurs at later stages of infection (Wang et al. 2012). It seems that at early stages of infection the phytosterol biosynthesis is inhibited to reroute the substrates to carotenoid and tocopherol biosynthesis pathways, which may be involved in photosystem protection during the oxidative burst (Munné-Bosch 2005; Rastogi et al. 2013). However, the levels of neither tocopherols nor carotenoids change during the studied early hours of infection. It could be argued that those compounds are immediately utilized during the oxidative burst in response to the infection (Atanasova-Penichon et al. 2016), but the discrepancy between gene expression and compound levels seemed too large. Carotenoids, despite being potent, fat-soluble antioxidants, constitute substrates for a number of derivatives, including ABA. Therefore we turned towards carotenoid cleavage pathways in order to determine the fate of synthesized carotenoids. The NCEDs were strongly activated, together with other genes leading to ABA synthesis. Correspondingly, the levels of accumulated ABA were strongly increasing continuously during the first 36 h. The observed activation of NCED genes upon infection corresponds with an another report where NCED2, NCED3, and NCED5 were induced, and ABA accumulated strongly following compatible P. syringae infection of A. thaliana (Fan et al. 2009). On the other hand, Liu et al. (2015) showed that NCED3 and NCED5 were repressed by WRKY33, a transcription factor activated in response to Botrytis cinerea, but this mechanism is likely not to occur during the flax response to F. oxysporum.
While the role of ABA in abiotic stresses (in particular drought) is quite well understood, its function in pathogen response is not so well established. ABA is thought to modulate processes that impact resistance, including stomatal closure, defense gene expression, and ROS production/scavenging, indicating an extensive role of ABA in defense. For example, ABA was reported to increase plant resistance to pathogen attack, mostly through remodeling of the cell wall, including changing callose deposition (Zeyen et al. 2002), leading to formation of plant papillae that hinder pathogen infection, and induction of phenylalanine ammonia lyase (PAL) at the transcriptional level in soybean plants (P. sojae) (Ryerson et al. 1993). Recent models suggest that ABA may have different roles that vary depending on the type of tissue infected, stage of disease, the pathogen infection strategy, and/or the pathogen (Maksimov 2009). Current understanding is that the role of ABA in defense cannot be generalized as it appears to have a pathogen- and context-dependent role (Cao et al. 2011; Lim and Lee 2015).
One of the first defense mechanisms of a plant to pathogen infection is the formation of papillae. Their role is to slow down the spread of a pathogen and to buy the time for activating more complex and more efficient mechanisms. The papillae are composed mainly of callose, but also of phenolic compounds, reactive oxygen species or thionins with antimicrobial activity (Voigt 2014). It was shown that ABA is a positive regulator of papilla formation through influencing callose deposition. However, the process is not fully understood; in B. cinerea it is based on post-transcriptional regulation of genes involved in callose synthesis (García-Andrade et al. 2011). Callose deposition can be regulated by numerous factors, including ABA (Chen and Kim 2009). In our study, together with the infection progress, we observed an increased expression of the PMR4 gene (encoding callose synthase 5). The level of expression of the gene was connected with the callose content in flax seedlings, which increased to its maximum at 48 hpi. In-silico analysis of the flax PMR4 gene promoter revealed the presence of cis-regulatory sequences responsive to ABA—DRE2COREZMRAB17, ABRELATERD1, ABRERATCA. This is supported by our preliminary data on the comparison of flax transcriptomes after 100 μM ABA treatment and after pathogen infection, in which similar groups of PR genes and genes connected with pathogenesis [e.g., PR3, PR4—chitinases, PR2—1,3-β-glucanases, PR5—TLP (thaumatin-like protein), gene encoding transcription factor TGA1] are similarly expressed (Suppl. Table S3) with the correlation coefficient of 0.45 (P < 0.005). Callose synthase genes are evolved in higher plants for the synthesis of callose in different tissues and in response to different physiological and developmental signals (Liu et al. 2015). Ellinger and Voigt (2014) reported that the expression of AtCalS genes is regulated in a tissue-specific manner. While in our experiments we investigated the whole plant response, it would be of interest to examine callose synthesis in various tissue types upon F. oxysporum infection in future. Moreover, the callose synthase genes were induced by salicylic acid (SA) treatment (Dong et al. 2008), and thus further investigations are required whether the gene is activated directly due to ABRE presence in the promoter, through a more complex phytohormonal interplay, and whether the mechanisms might be tissue specific. Similarly, chitinase, another PR protein induced by pathogen infection, is expressed in a tissue-dependent manner (Cao and Tan 2019). As we described previously (Boba et al. 2018), exogenous ABA application in flax leads to increased expression of the chitinase gene, which also possesses ABA-responsive cis-regulatory elements. Transgenic flax plants with an elevated ABA level showed increased resistance to fusariosis (Boba et al. 2018), which may be associated with higher expression of the chitinase gene. This was also shown in wheat, where a more resistant cultivar, ‘Sumai 3’, was characterized by twofold higher levels of ABA, 7′-OH ABA and ABA glucose ester, as well as SA, JA, and JA-isoleucine in relation to the FHB-susceptible Fielder (Buhrow et al. 2016). Regardless of the direct cause, our staining experiments of plant roots infected with F. oxysporum showed slower pathogen spread when plants were treated with ABA prior to the infection, compared to the non-treated control as soon as 4 hpi, which was confirmed by RT-PCR, indicating the involvement of ABA in the early plant response.
Time-course gene expression and metabolite measurements suggest a terpenoid pathway induction at the early stage of plant infection. It seems to occur even earlier than the phenylpropanoid pathway activation (Kostyn et al. 2012). This might indicate that the terpenoid and phenylpropanoid pathways are activated upon infection and may be somehow related, but the primary response originates from the terpenoid pathway. The relation between the two pathways may result from ABA-related recruitment of transcription factors that activate the expression of genes of the phenylpropanoid pathway. There are reports of the involvement of WRKY factors in the regulation of biosynthesis of several stress-induced secondary metabolites such as lignin (Guillaumie et al. 2009; Levée et al. 2009), tannin in the seed coat (Johnson et al. 2002), or alkaloids (Kato et al. 2007). AtWRKY23 regulates the production of flavonols in A. thaliana (Grunewald et al. 2012), while in Catharanthus roseus a number of WRKYs are involved in phytoalexin biosynthesis in response to biotic stress (Schluttenhofer et al. 2014). Moreover, LuWRKY36 was shown to play a role in the regulation of lignan biosynthesis in response to ABA and Fusarium elicitation through the binding of W boxes found in the promoter of LuPLR1 (Markulin et al. 2019). Our preliminary data on the comparison of flax transcriptomes after 100 μM ABA treatment and after pathogen infection indicate an upregulation of three WRKY transcription factors.
Abbreviations
- hpi:
-
Hours post infection
- JA:
-
Jasmonic acid
- MEP pathway:
-
Methylerythritol 4-phosphate pathway (non-mevalonate)
- PR:
-
Pathogenesis-related
References
Adie BAT, Pérez-Pérez J, Pérez-Pérez MM et al (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19(5):1665–1681
Atanasova-Penichon V, Barreau C, Richard-Forget F (2016) Antioxidant secondary metabolites in cereals: potential involvement in resistance to Fusarium and mycotoxin accumulation. Front Microbiol 7:566–566
Boba A, Kulma A, Kostyn K et al (2011) The influence of carotenoid biosynthesis modification on the Fusarium culmorum and Fusarium oxysporum resistance in flax. Physiol Mol Plant Pathol 76(1):39–47
Boba A, Kostyn K, Kostyn A et al (2016) Methyl salicylate level increase in flax after Fusarium oxysporum infection is associated with phenylpropanoid pathway activation. Front Plant Sci 7:1951
Boba A, Kostyn K, Preisner M et al (2018) Expression of heterologous lycopene β-cyclase gene in flax can cause silencing of its endogenous counterpart by changes in gene-body methylation and in ABA homeostasis mechanism. Plant Physiol Biochem 127:143–151
Buhrow LM, Cram D, Tulpan D et al (2016) Exogenous abscisic acid and gibberellic acid elicit opposing effects on Fusarium graminearum infection in wheat. Phytopathology 106(9):986–996
Cao J, Tan X (2019) Comprehensive analysis of the chitinase family genes in tomato (Solanum lycopersicum). Plants 8(3):52
Cao FY, Yoshioka K, Desveaux D (2011) The roles of ABA in plant–pathogen interactions. J Plant Res 124(4):489–499
Chen X-Y, Kim J-Y (2009) Callose synthesis in higher plants. Plant Signal Behav 4(6):489–492
Cole SJ, Yoon AJ, Faull KF et al (2014) Host perception of jasmonates promotes infection by Fusarium oxysporum formae speciales that produce isoleucine- and leucine-conjugated jasmonates. Mol Plant Pathol 15(6):589–600
De Gara L, de Pinto MC, Tommasi F (2003) The antioxidant systems vis-à-vis reactive oxygen species during plant–pathogen interaction. Plant Physiol Biochem 41(10):863–870
Di X, Takken FLW, Tintor N (2016) How phytohormones shape interactions between plants and the soil-borne fungus Fusarium oxysporum. Front Plant Sci 7:170
Di X, Gomila J, Takken FLW (2017) Involvement of salicylic acid, ethylene and jasmonic acid signalling pathways in the susceptibility of tomato to Fusarium oxysporum. Mol Plant Pathol 18(7):1024–1035
Dong X, Hong Z, Chatterjee J et al (2008) Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta 229(1):87–98
Fan J, Hill L, Crooks C et al (2009) Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol 150(4):1750–1761
Finkelstein R (2013) Abscisic acid synthesis and response. Arabidopsis Book Am Soc Plant Biol 11:e0166
García-Andrade J, Ramírez V, Flors V et al (2011) Arabidopsis OCP3 mutant reveals a mechanism linking ABA and JA to pathogen-induced callose deposition. Plant J 67(5):783–794
Gomez-Porras JL, Riano-Pachon DM, Dreyer I et al (2007) Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genom 8:260
Gonzalez-Candelas L, Alamar S, Sanchez-Torres P et al (2010) A transcriptomic approach highlights induction of secondary metabolism in citrus fruit in response to Penicillium digitatum infection. BMC Plant Biol 10:194
Grunewald W, De Smet I, Lewis DR et al (2012) Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci USA 109(5):1554–1559
Guillaumie S, Mzid R, Méchin V et al (2009) The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol Biol 72(1):215
Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14(6):1359
Karlovsky P (2011) Biological detoxification of the mycotoxin deoxynivalenol and its use in genetically engineered crops and feed additives. Appl Microbiol Biotechnol 91(3):491–504
Kato N, Dubouzet E, Kokabu Y et al (2007) Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol 48(1):8–18
Knight NL, Sutherland MW (2011) A rapid differential staining technique for Fusarium pseudograminearum in cereal tissues during crown rot infections. Plant Pathol 60(6):1140–1143
Kostyn K, Czemplik M, Kulma A et al (2012) Genes of phenylpropanoid pathway are activated in early response to Fusarium attack in flax plants. Plant Sci 190:103–115
Kroes GMLW, Baayen RP, Lange W (1998) Histology of root rot of flax seedlings (Linum usitatissimum) infected by Fusarium oxysporum f.sp. lini. Eur J Plant Pathol 104(7):725–736
Levée V, Major I, Levasseur C et al (2009) Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol 184(1):48–70
Li C, Zuo C, Deng G et al (2013) Contamination of bananas with beauvericin and fusaric acid produced by Fusarium oxysporum f. sp. cubense. PLoS One 8(7):e70226
Lim CW, Lee SC (2015) Arabidopsis abscisic acid receptors play an important role in disease resistance. Plant Mol Biol 88(3):313–324
Liu S, Kracher B, Ziegler J et al (2015) Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. eLife 4:e07295–e07295
Lushchak VI, Semchuk NM (2012) Tocopherol biosynthesis: chemistry, regulation and effects of environmental factors. Acta Physiol Plant 34(5):1607–1628
Maksimov IV (2009) Abscisic acid in the plants-pathogen interaction. Russ J Plant Physiol 56(6):742–752
Markulin L, Corbin C, Renouard S, et al. (2019) Characterization of LuWRKY36, a flax transcription factor promoting secoisolariciresinol biosynthesis in response to Fusarium oxysporum elicitors in Linum usitatissimum L. hairy roots. Planta 250(1):347–366
Michielse CB, Rep M (2009) Pathogen profile update: Fusarium oxysporum. Mol Plant Pathol 10(3):311–324
Mittler R, Blumwald E (2015) The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27(1):64–70
Munné-Bosch S (2005) The role of α-tocopherol in plant stress tolerance. J Plant Physiol 162(7):743–748
Nisar N, Li L, Lu S et al (2015) Carotenoid metabolism in plants. Mol Plant 8(1):68–82
Radhakrishnan R, Pae SB, Shim KB et al (2013) Penicillium sp. mitigates Fusarium-induced biotic stress in sesame plants. Biotechnol Lett 35(7):1073–1078
Rastogi A, Yadav D, Szymańska R et al (2013) Singlet oxygen scavenging activity of tocopherol and plastochromanol in Arabidopsis thaliana: Relevance to photooxidative stress. Plant Cell Environ 37(2):392–401
Ryerson E, Li A, Young JP et al (1993) Changes in abscisic acid levels in bean leaves during the initial stages of host and nonhost reactions to rust fungi. Physiol Mol Plant Pathol 43(4):265–273
Sanchez-Vallet A, Lopez G, Ramos B et al (2012) Disruption of abscisic acid signaling constitutively activates Arabidopsis resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant Physiol 160(4):2109–2124
Schluttenhofer C, Pattanaik S, Patra B et al (2014) Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement in jasmonate signaling. BMC Genom 15(1):502
Shi W, Tan Y, Wang S et al (2017) Mycotoxigenic potentials of Fusarium species in various culture matrices revealed by mycotoxin profiling. Toxins 9(1):6
Sonawane PD, Pollier J, Panda S et al (2016) Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat Plants 3:16205
Song W, Ma X, Tan H et al (2011) Abscisic acid enhances resistance to Alternaria solani in tomato seedlings. Plant Physiol Biochem 49(7):693–700
Thatcher L, Manners J, Kazan K (2009) Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J 58(6):927–939
Ton J, Flors V, Mauch-Mani B (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci 14(6):310–317
Voigt CA (2014) Callose-mediated resistance to pathogenic intruders in plant defense-related papillae. Front Plant Sci 5:168
Wagacha JM, Muthomi JW (2007) Fusarium culmorum: Infection process, mechanisms of mycotoxin production and their role in pathogenesis in wheat. Crop Protect 26(7):877–885
Wang K, Senthil-Kumar M, Ryu C-M et al (2012) Phytosterols play a key role in plant innate immunity against bacterial pathogens by regulating nutrient efflux into the apoplast. Plant Physiol 158(4):1789–1802
Wang Y, Tao X, Tang X-M et al (2013) Comparative transcriptome analysis of tomato (Solanum lycopersicum) in response to exogenous abscisic acid. BMC Genom 14(1):841
Wojtasik W, Kulma A, Kostyn K et al (2011) The changes in pectin metabolism in flax infected with Fusarium. Plant Physiol Biochem 49(8):862–872
Zeyen RJ, Kruger WM, Lyngkjær MF et al (2002) Differential effects of d-mannose and 2-deoxy-d-glucose on attempted powdery mildew fungal infection of inappropriate and appropriate G ramineae. Physiol Mol Plant Pathol 61(6):315–323
Acknowledgements
This research project was funded by the Polish National Science Centre (NCN), Grant Nos. NCN 2018/29/B/NZ9/00288, 2011/01/B/NZ1/00217 and 2014/15/B/NZ9/00470.
Author information
Authors and Affiliations
Contributions
AB performed infection tests and gene expression analysis, and participated in writing of the manuscript; KK performed metabolite analysis and wrote the manuscript; BK performed transcriptome analysis; WW participated in metabolite analysis, and prepared figures; MP participated in metabolite analysis; AP performed sterol analysis; EMG performed staining experiments; DL participated in sterol analysis; BD participated in writing the manuscript; JS coordinated the study, and participated in data analysis; AK planned the experiments and participated in writing of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Boba, A., Kostyn, K., Kozak, B. et al. Fusarium oxysporum infection activates the plastidial branch of the terpenoid biosynthesis pathway in flax, leading to increased ABA synthesis. Planta 251, 50 (2020). https://doi.org/10.1007/s00425-020-03339-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-020-03339-9