Abstract
Main conclusion
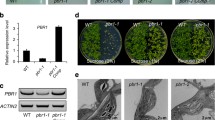
Chloroplasts deficient in the major chloroplast nucleoid-associated protein WHIRLY1 have an enhanced ratio of LHCs to reaction centers, indicating that WHIRLY1 is required for a coordinate assembly of the photosynthetic apparatus during chloroplast development.
Abstract
Chloroplast development was found to be delayed in barley plants with an RNAi-mediated knockdown of WHIRLY1 encoding a major nucleoid-associated protein of chloroplasts. The plastids of WHIRLY1 deficient plants had a reduced ribosome content. Accordingly, plastid-encoded proteins of the photosynthetic apparatus showed delayed accumulation during chloroplast development coinciding with a delayed increase in photosystem II efficiency measured by chlorophyll fluorescence. In contrast, light harvesting complex proteins being encoded in the nucleus had a high abundance as in the wild type. The unbalanced assembly of the proteins of the photosynthetic apparatus in WHIRLY1-deficient plants coincided with the enhanced contents of chlorophyll b and xanthophylls. The lack of coordination was most obvious at the early stages of development. Overaccumulation of LHC proteins in comparison to reaction center proteins at the early stages of chloroplast development did not correlate with enhanced expression levels of the corresponding genes in the nucleus. This work revealed that WHIRLY1 does not influence LHC abundance at the transcriptional level. Rather, WHIRLY1 in association with nucleoids might play a structural role for both the assembly of ribosomes and the complexes of the photosynthetic apparatus.






Similar content being viewed by others
References
Anderson JM, Chow WS, Goodchild DJ (1988) Thylakoid membrane organization in sun shade acclimation. Aust J Plant Phys 15:11–26
Baumgartner BJ, Rapp JC, Mullet JE (1993) Plastid genes encoding the transcription translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development—evidence for selective stabilization of psbA messenger RNA. Plant Physiol 101:781–791
Belcher S, Williams-Carrier R, Stiffler N, Barkan A (2015) Large-scale genetic analysis of chloroplast biogenesis in maize. Biochim Biophys Acta Bioenerg 1847:1004–1016
Bohne AV (2014) The nucleoid as a site of rRNA processing and ribosome assembly. Front Plant Sci 5:257
Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV (2015) Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim Biophys Acta Bioenerg 1847:761–769
Bradbeer J, Atkinson Y, Börner T, Hagemann R (1979) Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesized RNA. Nature 279:816–817
Caffarri S, Tibiletti T, Jennings RC, Santabarbara S (2014) A comparison between plant photosystem I and photosystem II architecture and functioning. Curr Protein Pept Sci 15:296–331
Cappadocia L, Parent JS, Zampini E, Lepage E, Sygusch J, Brisson N (2012) A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage. Nucleic Acids Res 40:258–269
Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol 67:25–53
Crepin A, Caffarri S (2018) Functions and evolution of Lhcb isoforms composing LHCII, the major light harvesting complex of photosystem II of green eukaryotic organisms. Curr Protein Pept Sci 19:699–713
Desveaux D, Després C, Joyeux A, Subramaniam R, Brisson N (2000) PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12:279–290
Desveaux D, Subramaniam R, Deprés C, Mess J-N, Lévesque C, Fobert PR, Dangl J, Brisson N (2004) A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopis. Dev Cell 6:229–240
Desveaux D, Maréchal A, Brisson N (2005) Whirly transcription factors: defense gene regulation and beyond. Trends Plant Sci 10:95–102
Dong T, Park Y, Hwang I (2015) Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling. In: Guilfoyle T, Hagen G (eds) Plant hormone signalling, essays in biochemistry. Portland Press LTD, London, pp 29–48
Fling S, Gregerson D (1986) Peptide and protein molecular weight determination by electrophoresis using high-molarity tris buffer without urea. Anal Biochem 155:83–88
Grabowski E, Miao Y, Mulisch M, Krupinska K (2008) Single-stranded DNA binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol 147:1800–1804
Green BR (2011) Chloroplast genomes of photosynthetic eukaryotes. Plant J 66:34–44
Gregersen PL, Culetic A, Boschian L, Krupinska K (2013) Plant senescence and crop productivity. Plant Mol Biol 82:603–622
Hernandez-Verdeja T, Strand A (2018) Retrograde signals navigate the path to chloroplast development. Plant Physiol 176:967–976
Hess W, Müller A, Nagy F, Börner T (1994) Ribosome-deficient plastids affect transcription of light-induced nuclear genes: genetic evidence for a plastid-derived signal. Mol Gen Genet 242:305–312
Humbeck K, Quast S, Krupinska K (1996) Functional and molecular changes in the photosynthetic apparatus during senescence of flag leaves from field-grown barley plants. Plant Cell Environ 19:337–344
Isemer R (2013) Die WHIRLY-Proteine von Arabidopsis thaliana. Dissertation, Naturwissenschaftliche Fakultät der Christian-Albrechts-Universität Kiel, Germany
Isemer R, Mulisch M, Schäfer A, Kirchner S, Koop H-U, Krupinska K (2012) Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett 586:85–88
Janack B, Sosoi P, Krupinska K, Humbeck K (2016) Knockdown of WHIRLY1 affects drought stress-induced leaf senescence and histone modifications of the senescence-associated gene HvS40. Plants 5:37
Jensen P, Bassi R, Boekema E, Dekker J, Jansson S, Leister D, Robinson C, Scheller H (2007) Structure, function and regulation of plant photosystem I. Biochim Biophys Acta 1767:335–352
Koussevitzky S, Nott A, Mockler T, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316:715–719
Krupinska K, Melonek J, Krause K (2013) New insights into plastid nucleoid structure and functionality. Planta 237:653–664
Krupinska K, Oetke S, Desel C, Mulisch M, Schäfer A, Hollmann J, Kumlehn J, Hensel G (2014) WHIRLY1 is a major organizer of chloroplast nucleoids. Front Plant Sci 5:432
Kucharewicz W, Distelfeld A, Bilger W, Muller M, Munne-Bosch S, Hensel G, Krupinska K (2017) Acceleration of leaf senescence is slowed down in transgenic barley plants deficient in the DNA/RNA-binding protein WHIRLY1. J Exp Bot 68:983–996
Leister D, Romani I, Mittermayr L, Paieri F, Fenino E, Kleine T (2014) Identification of target genes and transcription factors implicated in translation-dependent retrograde signaling in Arabidopsis. Mol Plant 7:1228–1247
Luo T, Luo S, Araujo WL, Schlicke H, Rothbart M, Yu J, Fan TT, Fernie AR, Grimm B, Luo MZ (2013) Virus-induced gene silencing of pea CHLI and CHLD affects tetrapyrrole biosynthesis, chloroplast development and the primary metabolic network. Plant Physiol Biochem 65:17–26
Maréchal A, Parent JS, Veronneau-Lafortune F, Joyeux A, Lang BF, Brisson N (2009) Whirly proteins maintain plastid genome stability in Arabidopsis. Proc Natl Acad Sci USA 106:14693–14698
Marjanac G, Karimi M, Naudts M, Beeckman T, Depicker A, De Buck S (2009) Gene silencing induced by hairpin or inverted repeated sense transgenes varies among promoters and cell types. New Phytol 184:851–864
Melonek J, Mulisch M, Schmitz-Linneweber C, Grabowski E, Hensel G, Krupinska K (2010) Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta 232:471–481
Melonek J, Oetke S, Krupinska K (2016) Multifunctionality of plastid nucleoids as revealed by proteome analyses. Biochim Biophys Acta 1864:1016–1038
Nakagawara E, Sakuraba Y, Yamasato A, Tanaka R, Tanaka A (2007) Clp protease controls chlorophyll b synthesis by regulating the level of chlorophyllide a oxygenase. Plant J 49:800–809
Niinemets Ü, Bilger W, Kull O, Tenhunen JD (1998) Acclimation to high irradiance in temperate deciduous trees in the field: changes in xanthophyll cycle pool size and in photosynthetic capacity along a canopy light gradient. Plant Cell Environ 21:1205–1218
Nishimura K, Kata Y, Sakamoto W (2016) Chloroplast proteases: updates on proteolysis within and across suborganellar compartments. Plant Physiol 171:2280–2293
Pfalz J, Liere K, Kandlbinder A, Dietz K-J, Oelmüller R (2006) pTAC2-6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18:176–197
Pfalz J, Liebers M, Hirth M, Grubler B, Holtzegel U, Schroter Y, Dietzel L, Pfannschmidt T (2012) Environmental control of plant nuclear gene expression by chloroplast redox signals. Front Plant Sci 3:257
Pfannschmidt T (2010) Plastidial retrograde signalling—a true “plastid factor” or just metabolite signatures? Trends Plant Sci 15:427–435
Piechulla B (1999) Circadian expression of the light-harvesting complex protein genes in plants. Chronobiol Int 16:115–128
Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A (2008) A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res 36:5152–5165
Race HL, Herrmann RG, Martin W (1999) Why have organelles retained genomes? Trends Genet 15:364–370
Rapacz M, Stepien A, Skorupa K (2012) Internal standards for quantitative RT-PCR studies of gene expression under drought treatment in barley (Hordeum vulgare L.): the effects of developmental stage and leaf age. Acta Physiol Plant 34:1723–1733
Sakamoto W, Uno Y, Zhang Q, Miura E, Kato Y, Sodmergen (2009) Arrested differentiation of proplastids into chloroplasts in variegated leaves characterized by plastid ultrastructure and nucleoid morphology. Plant Cell Physiol 50:2069–2083
Sakuraba Y, Balazadeh S, Tanaka R, Mueller-Roeber B, Tanaka A (2012) Overproduction of chl b retards senescence through transcriptional reprogramming in Arabidopsis. Plant Cell Physiol 53:505–517
Schöttler MA, Toth SZ (2014) Photosynthetic complex stoichiometry dynamics in higher plants: environmental acclimation and photosynthetic flux control. Front Plant Sci 5:188
Seo M, Koshiba T (2011) Transport of ABA from the site of biosynthesis to the site of action. J Plant Res 124:501–507
Swida-Barteczka A, Krieger-Liszkay A, Bilger W, Voigt U, Hensel G, Szweykowska-Kulinska Z, Krupinska K (2018) The plastid-nucleus located DNA/RNA binding protein WHIRLY1 regulates microRNA-levels during stress in barley (Hordeum vulgare L.). RNA Biol. https://doi.org/10.1080/15476286.2018.1481695
Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7:1105–1120
Vallon O, Hoyer-Hansen G, Simpson DJ (1987) Photosystem II and cytochrome b559 in the stroma lamellae of barley chloroplasts. Carlsberg Res Commun 52:405–421
Voitsekhovskaja OV, Tyutereva EV (2015) Chlorophyll b in angiosperms: functions in photosynthesis, signaling and ontogenetic regulation. J Plant Physiol 189:51–64
Wobbe L, Bassi R, Kruse O (2016) Multi-level light capture control in plants and green algae. Trends Plant Sci 21:55–68
Wu GZ, Chalvin C, Hoelscher M, Meyer EH, Wu XN, Bock R (2018) Control of retrograde signaling by rapid turnover of genomes uncoupled1. Plant Physiol 176:2472–2495
Xu YH, Liu R, Yan L, Liu ZQ, Jiang SC, Shen YY, Wang XF, Zhang DP (2012) Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J Exp Bot 63:1095–1106
Yoo HH, Kwon C, Lee MM, Chung IK (2007) Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J 49:442–451
Yu F, Fu AG, Aluru M, Park S, Xu Y, Liu HY, Liu XY, Foudree A, Nambogga M, Rodermel S (2007) Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ 30:350–365
Zhelyazkova P, Sharma CM, Forstner KU, Liere K, Vogel J, Börner T (2012) The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24:123–136
Acknowledgements
We acknowledge the technical assistance of Ulrike Voigt and Jens Hermann (Institute of Botany, CAU Kiel, Germany). Luca Boschian is thanked for discussion and Svenja Oetke is thanked for critical reading of the manuscript. This research was supported by the German Science Foundation (KR 1350/19-1).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Krupinska, K., Braun, S., Nia, M.S. et al. The nucleoid-associated protein WHIRLY1 is required for the coordinate assembly of plastid and nucleus-encoded proteins during chloroplast development. Planta 249, 1337–1347 (2019). https://doi.org/10.1007/s00425-018-03085-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-018-03085-z