Abstract
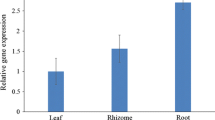
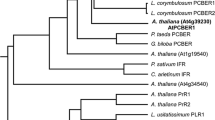
Some flavonoids are considered as beneficial compounds because they exhibit anticancer or antioxidant activity. In higher plants, flavonoids are secondary metabolites that are derived from phenylpropanoid biosynthetic pathway. A large number of phenylpropanoids are generated from p-coumaric acid, which is a derivative of the primary metabolite, phenylalanine. The first two steps in the phenylpropanoid biosynthetic pathway are catalyzed by phenylalanine ammonia-lyase and cinnamate 4-hydroxylase, and the coupling of these two enzymes forms a rate-limiting step in the pathway. For the generation of p-coumaric acid, the conversion from phenylalanine to p-coumaric acid that is catalyzed by two enzymes can be theoretically performed by a single enzyme, tyrosine ammonia-lyase (TAL) that catalyzes the conversion of tyrosine to p-coumaric acid in certain bacteria. To modify the p-coumaric acid pathway in plants, we isolated a gene encoding TAL from a photosynthetic bacterium, Rhodobacter sphaeroides, and introduced the gene (RsTAL) in Arabidopsis thaliana. Analysis of metabolites revealed that the ectopic over-expression of RsTAL leads to higher accumulation of anthocyanins in transgenic 5-day-old seedlings. On the other hand, 21-day-old seedlings of plants expressing RsTAL showed accumulation of higher amount of quercetin glycosides, sinapoyl and p-coumaroyl derivatives than control. These results indicate that ectopic expression of the RsTAL gene in Arabidopsis enhanced the metabolic flux into the phenylpropanoid pathway and resulted in increased accumulation of flavonoids and phenylpropanoids.




Similar content being viewed by others
Abbreviations
- C4H:
-
Cinnamate 4-hydroxylase
- 4CL:
-
4-Coumarate:CoA ligase
- ORF:
-
Open reading frame
- PAL:
-
Phenylalanine ammonia-lyase
- PCR:
-
Polymerase chain reaction
- RsTAL :
-
Rhodobacter sphaeroides TAL
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- TAL:
-
Tyrosine ammonia-lyase
References
Achnine L, Blancaflor EB, Rasmussen S, Dixon RA (2004) Colocalization of l-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16:3098–3109
Appert C, Logemann E, Hahlbrock K, Schmid J, Amrhein N (1994) Structural and catalytic properties of the four phenylalanine ammonia-lyase isoenzymes from parsley (Petroselinum crispum Nym.). Eur J Biochem 225:491–499
Bate NJ, Orr J, Ni W, Meromi A, Nadler-Hassar T, Doerner PW, Dixon RA, Lamb CJ, Elkind Y (1994) Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc Natl Acad Sci USA 91:7608–7612
Blount JW, Korth KL, Masoud SA, Rasmussen S, Lamb C, Dixon RA (2000) Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol 122:107–116
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546
Bolwell GP, Cramer CL, Lamb CJ, Schuch W, Dixon RA (1986) l-Phenylalanine ammonia-lyase from Phaseolus vulgaris: modulation of the levels of active enzyme by trans-cinnamic acid. Planta 169:97–107
Bolwell GP, Mavandad M, Millar DJ, Edwards KJ, Schuch W, Dixon RA (1988) Inhibition of mRNA levels and activities by trans-cinnamic acid in elicitor-induced bean cells. Phytochemistry 27:2109–2117
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2393
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cochrane FC, Davin LB, Lewis NG (2004) The Arabidopsis phenylalanine ammonia lyase gene family: kinetic characterization of the four PAL isoforms. Phytochemistry 65:1557–1564
Dixon RA (2004) Phytoestrogens. Annu Rev Plant Biol 55:225–261
Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085–1097
Ehlting J, Büttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E (1999) Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19:9–20
Elkind Y, Edwards R, Mavandad M, Hedrick SA, Ribak O, Dixon RA, Lamb CJ (1990) Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proc Natl Acad Sci USA 87:9057–9061
Fischer R, Budde I, Hain R (1997) Stilbene synthase gene expression causes changes in flower colour and male sterility in tobacco. Plant J 11:489–498
Hahlbrock K, Knobloch KH, Kreuzaler F (1976) Coordinated induction and subsequent activity changes of two groups of metabolically interrelated enzymes: Light induced synthesis of flavonoid glycosides in cell suspension cultures of Petroselinum hortense. Eur J Biochem 61:199–206
Höfgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16:9877
Howles PA, Sewalt VJH, Paiva NL, Elkind Y, Bate NJ, Lamb C, Dixon RA (1996) Overexpression of l-phenylalanine ammonia-lyase in transgenic tobacco plants reveals control points for flux into phenylpropanoid biosynthesis. Plant Physiol 112:1617–1624
Jorrin J, Dixon RA (1990) Stress responses in alfalfa (Medicago sativa L.): II. Purification, characterization, and induction of phenylalanine ammonia-lyase isoforms from elicitor-treated cell suspension cultures. Plant Physiol 92:447–455
Kanno T, Komatsu A, Kasai K, Dubouzet JG, Sakurai M, Ikejiri-Kanno Y, Wakasa K, Tozawa Y (2005) Structure-based in vitro engineering of the anthranilate synthase, a metabolic key enzyme in the plant tryptophan pathway. Plant Physiol 138:2260–2268
Kort R, Phillips-Jones MK, van Aalten DMF, Haker A, Hoffer SM, Hellingwerf KJ, Crielaard W (1998) Sequence, chromophore extraction and 3-D model of the photoactive yellow protein from Rhodobacter sphaeroides. Biochim Biophys Acta 1385:1–6
Kyndt JA, Meyer TE, Cusanovich MA, Van Beeumen JJ (2002) Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Lett 512:240–244
Matsuda F, Yonekura-Sakakibara K, Niida R, Kuromori T, Shinozaki K, Saito K (2009) MS/MS spectral tag (MS2T)-based annotation of non-targeted profile of plant secondary metabolites. Plant J 57:555–577
Mavandad M, Edwards R, Liang X, Lamb CJ, Dixon RA (1990) Effects of trans-cinnamic acid on expression of the bean phenylalanine ammonia-lyase gene family. Plant Physiol 94:671–680
Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138:1083–1096
Millar DJ, Long M, Donovan G, Fraser PD, Boudet AM, Danoun S, Bramley PM, Bolwell GP (2007) Introduction of sense constructs of cinnamate 4-hydroxylase (CYP73A24) in transgenic tomato plants shows opposite effects on flux into stem lignin and fruit flavonoids. Phytochemistry 68:1497–1509
Pietta PG (2000) Flavonoids as antioxidants. J Nat Prod 63:1035–1042
Ren W, Qiao Z, Wang H, Zhu L, Zhang L (2003) Flavonoids: promising anticancer agents. Med Res Rev 23:519–534
Rösler J, Krekel F, Amrhein N, Schmid J (1997) Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol 113:175–179
Shadle GL, Wesley SV, Korth KL, Chen F, Lamb C, Dixon RA (2003) Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of l-phenylalanine ammonia-lyase. Phytochemistry 64:153–161
Sharma SB, Dixon RA (2005) Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana. Plant J 44:62–75
Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54:733–749
Tohge T, Nishiyama Y, Yokota-Hirai M, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, Noji M, Yamazaki M, Saito K (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42:218–235
Tozawa Y, Tanaka K, Takahashi H, Wakasa K (1998) Nuclear encoding of a plastid σ factor in rice and its tissue- and light-dependent expression. Nucleic Acids Res 26:415–419
Watts KT, Lee PC, Schmidt-Dannert C (2004) Exploring recombinant flavonoid biosynthesis in metabolically engineered Escherichia coli. Chembiochem 5:500–507
Williams CA, Grayer RJ (2004) Anthocyanins and other flavonoids. Nat Prod Rep 21:539–573
Williams RJ, Spencer JP, Rice-Evans C (2004) Flavonoids: antioxidants or signaling molecules? Free Rad Biol Med 36:838–849
Xie DY, Sharma SB, Wright E, Wang ZY, Dixon RA (2006) Metabolic engineering of proanthocyanidins through co-expression of anthocyanidin reductase and the PAP1 MYB transcription factor. Plant J 45:895–907
Xue Z, McCluskey M, Cantera K, Sariaslani FS, Huang L (2007) Identification, characterization and functional expression of a tyrosine ammonia-lyase and its mutants from the photosynthetic bacterium Rhodobacter sphaeroides. J Ind Microbiol Biotechnol 34:599–604
Ylstra B, Busscher J, Franken J, Hollman PCH, Mol JNM, van Tunen AJ (1994) Flavonols and fertilization in Petunia hybrida: localization and mode of action during pollen tube growth. Plant J 6:201–212
Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K (2009) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene–metabolite correlations in Arabidopsis. Plant Cell 20:2160–2176
Yun CS, Yamamoto T, Nozawa A, Tozawa Y (2008) Expression of parsley flavone synthase I establishes the flavone biosynthetic pathway in Arabidopsis thaliana. Biosci Biotechnol Biochem 72:968–973
Author information
Authors and Affiliations
Corresponding author
Additional information
Y. Nishiyama and C. S. Yun contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nishiyama, Y., Yun, CS., Matsuda, F. et al. Expression of bacterial tyrosine ammonia-lyase creates a novel p-coumaric acid pathway in the biosynthesis of phenylpropanoids in Arabidopsis. Planta 232, 209–218 (2010). https://doi.org/10.1007/s00425-010-1166-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1166-1