Abstract
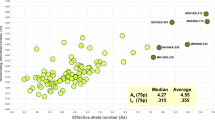
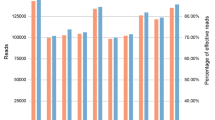
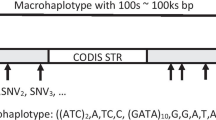
Mixture detection remains one of the major challenges within a forensic science context. In recent years, microhaplotypes were proposed to have great potential in mixture detection, although many of them are not as polymorphic as widely used short tandem repeat (STR) markers. In this study, 59 new highly polymorphic microhaplotypes were identified and sequenced with the NextSeq 500 Sequencer. Based on the whole 1000 Genomes Project dataset, the average effective number of alleles (Ae) of the 59 microhaplotypes was 5.44, and the Ae values of 36 of these microhaplotypes were > 5.00. Their genetic variations in 187 Han Chinese individuals were evaluated. The average allele coverage ratio (ACR) of heterozygotes across all loci was 0.96 ± 0.05. The number of observed alleles varied from 4 to 23, with an average of 8.8 alleles per microhaplotype locus. The average observed heterozygosity (Ho) of 59 loci was 0.77 ± 0.05, and the Ho values of 15 of these loci were > 0.80. All loci showed high polymorphisms with a discrimination power (DP) ranging from 0.80 to 0.97, and the average DP was 0.92 ± 0.03. The analysis of simulated mixtures demonstrated that the microhaplotypes reported here were highly polymorphic and performed well in forensic DNA mixture analysis. This study not only demonstrated the applicability of microhaplotypes in mixture analysis but also provided new choices for highly polymorphic microhaplotypes because after adding the markers identified here, the number of microhaplotypes with Ae values of > 4.00 will increase from ~ 50 to ~ 110.





Similar content being viewed by others
References
Yang J, Lin D, Deng C, Li Z, Pu Y, Yu Y, Li K, Li D, Chen P, Chen F (2019) The advances in DNA mixture interpretation. Forensic Sci Int 301:101–106. https://doi.org/10.1016/j.forsciint.2019.05.024
Ellegren H (2004) Microsatellites: simple sequences with complex evolution. Nat Rev Genet 5:435–445. https://doi.org/10.1038/nrg1348
Schlötterer C, Tautz D (1992) Slippage synthesis of simple sequence DNA. Nucleic Acids Res 20:211–215. https://doi.org/10.1093/nar/20.2.211
Levinson G, Gutman GA (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol 4:203–221. https://doi.org/10.1093/oxfordjournals.molbev.a040442
Oldoni F, Podini D (2019) Forensic molecular biomarkers for mixture analysis. Forensic Sci Int Genet 41:107–119. https://doi.org/10.1016/j.fsigen.2019.04.003
Kidd KK, Pakstis AJ, Speed WC, Lagace R, Wootton S, Chang J (2018) Selecting microhaplotypes optimized for different purposes. Electrophoresis 39:2815–2823. https://doi.org/10.1002/elps.201800092
Turchi C, Melchionda F, Pesaresi M, Tagliabracci A (2019) Evaluation of a microhaplotypes panel for forensic genetics using massive parallel sequencing technology. Forensic Sci Int Genet 41:120–127. https://doi.org/10.1016/j.fsigen.2019.04.009
Bennett L, Oldoni F, Long K, Cisana S, Madella K, Wootton S, Chang J, Hasegawa R, Lagacé R, Kidd KK, Podini D (2019) Mixture deconvolution by massively parallel sequencing of microhaplotypes. Int J Legal Med 133:719–729. https://doi.org/10.1007/s00414-019-02010-7
Zhu J, Lv M, Zhou N, Chen D, Jiang Y, Wang L, He W, Peng D, Li Z, Qu S, Wang Y, Wang H, Luo H, An G, Liang W, Zhang L (2019) Genotyping polymorphic microhaplotype markers through the Illumina® MiSeq platform for forensics. Forensic Sci Int Genet 39:1–7. https://doi.org/10.1016/j.fsigen.2018.11.005
Chen P, Deng C, Li Z, Pu Y, Yang J, Yu Y, Li K, Li D, Liang W, Zhang L, Chen F (2019) A microhaplotypes panel for massively parallel sequencing analysis of DNA mixtures. Forensic Sci Int Genet 40:140–149. https://doi.org/10.1016/j.fsigen.2019.02.018
Kidd KK, Pakstis AJ, Speed WC, Lagace R, Chang J, Wootton S, Ihuegbu N (2013) Microhaplotype loci are a powerful new type of forensic marker. Forensic Sci Int Genet Suppl Ser 4:e123–e124. https://doi.org/10.1016/j.fsigss.2013.10.063
Voskoboinik L, Motro U, Darvasi A (2018) Facilitating complex DNA mixture interpretation by sequencing highly polymorphic haplotypes. Forensic Sci Int Genet 35:136–140. https://doi.org/10.1016/j.fsigen.2018.05.001
Kidd KK, Speed WC (2015) Criteria for selecting microhaplotypes: mixture detection and deconvolution. Investig Genet 6:1. https://doi.org/10.1186/s13323-014-0018-3
van der Gaag KJ, de Leeuw RH, Laros JFJ, den Dunnen JT, de Knijff P (2018) Short hypervariable microhaplotypes: a novel set of very short high discriminating power loci without stutter artefacts. Forensic Sci Int Genet 35:169–175. https://doi.org/10.1016/j.fsigen.2018.05.008
de la Puente M, Phillips C, Xavier C, Amigo J, Carracedo A, Parson W, Lareu MV (2020) Building a custom large-scale panel of novel microhaplotypes for forensic identification using MiSeq and ion S5 massively parallel sequencing systems. Forensic Sci Int Genet 45:102213. https://doi.org/10.1016/j.fsigen.2019.102213
Pang J-B, Rao M, Chen Q-F, Ji AQ, Zhang C, Kang KL, Wu H, Ye J, Nie SJ, Wang L (2020) A 124-plex microhaplotype panel based on next-generation sequencing developed for forensic applications. Sci Rep 10:1945. https://doi.org/10.1038/s41598-020-58980-x
Kidd KK, Speed WC, Pakstis AJ, Podini DS, Lagacé R, Chang J, Wootton S, Haigh E, Soundararajan U (2017) Evaluating 130 microhaplotypes across a global set of 83 populations. Forensic Sci Int Genet 29:29–37. https://doi.org/10.1016/J.FSIGEN.2017.03.014
Qu N, Lin S, Gao Y, Liang H, Zhao H, Ou X (2020) A microhap panel for kinship analysis through massively parallel sequencing technology. Electrophoresis 41:246–253. https://doi.org/10.1002/elps.201900337
Sun S, Liu Y, Li J, Yang Z, Wen D, Liang W, Yan Y, Yu H, Cai J, Zha L (2020) Development and application of a nonbinary SNP-based microhaplotype panel for paternity testing involving close relatives. Forensic Sci Int Genet 46:102255. https://doi.org/10.1016/j.fsigen.2020.102255
Chen P, Yin C, Li Z, Pu Y, Yu Y, Zhao P, Chen D, Liang W, Zhang L, Chen F (2018) Evaluation of the microhaplotypes panel for DNA mixture analyses. Forensic Sci Int Genet 35:149–155. https://doi.org/10.1016/j.fsigen.2018.05.003
Kent WJ (2002) BLAT---the BLAST-like alignment tool. Genome Res 12:656–664. https://doi.org/10.1101/gr.229202
Kidd KK (2016) Proposed nomenclature for microhaplotypes. Hum Genomics 10:16. https://doi.org/10.1186/s40246-016-0078-y
Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, Zhang X, Wang J, Yang H, Fang L, Chen Q (2018) SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7:1–6. https://doi.org/10.1093/gigascience/gix120
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Perez J, Mitchell AA, Ducasse N, Tamariz J, Caragine T (2011) Estimating the number of contributors to two-, three-, and four-person mixtures containing DNA in high template and low template amounts. Croat Med J 52:314–326. https://doi.org/10.3325/cmj.2011.52.314
Bieber FR, Buckleton JS, Budowle B, Butler JM, Coble MD (2016) Evaluation of forensic DNA mixture evidence: protocol for evaluation, interpretation, and statistical calculations using the combined probability of inclusion. BMC Genet 17:125. https://doi.org/10.1186/s12863-016-0429-7
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106. https://doi.org/10.1111/j.1365-294X.2007.03089.x
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Young BA, Gettings KB, McCord B, Vallone PM (2019) Estimating number of contributors in massively parallel sequencing data of STR loci. Forensic Sci Int Genet 38:15–22. https://doi.org/10.1016/j.fsigen.2018.09.007
Oldoni F, Kidd KK, Podini D (2019) Microhaplotypes in forensic genetics. Forensic Sci Int Genet 38:54–69. https://doi.org/10.1016/j.fsigen.2018.09.009
Oldoni F, Bader D, Fantinato C, Wootton SC, Lagacé R, Kidd KK, Podini D (2020) A sequence-based 74plex microhaplotype assay for analysis of forensic DNA mixtures. Forensic Sci Int Genet 301:102367. https://doi.org/10.1016/j.fsigen.2020.102367
Yang L, Zhang X, Zhao L, Sun Y, Li J, Huang R, Hu L, Nie S (2018) Population data of 23 autosomal STR loci in the Chinese Han population from Guangdong Province in southern China. Int J Legal Med 132:133–135. https://doi.org/10.1007/s00414-017-1588-4
Li H, Peng D, Wang Y, Wu RG, Zhang YM, Li R, Sun HY (2018) Evaluation of genetic parameters of 23 autosomal STR loci in a Southern Chinese Han population. Ann Hum Biol 45:359–364. https://doi.org/10.1080/03014460.2018.1480731
Schirmer M, D’Amore R, Ijaz UZ, Hall N, Quince C (2016) Illumina error profiles: resolving fine-scale variation in metagenomic sequencing data. BMC Bioinformatics 17:125. https://doi.org/10.1186/s12859-016-0976-y
Funding
This work was supported by the National Natural Science Foundation of China (81801878, 81671873), and the Natural Science Foundation of Guangdong Province (2018A030313413).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Approval was obtained from the ethical committee of the Sun Yat-sen University (permit number: 2018-042).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Code availability
The in-house Perl script for selection of microhaplotypes is available upon request.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(XLSX 80 kb)
Rights and permissions
About this article
Cite this article
Wu, R., Li, H., Li, R. et al. Identification and sequencing of 59 highly polymorphic microhaplotypes for analysis of DNA mixtures. Int J Legal Med 135, 1137–1149 (2021). https://doi.org/10.1007/s00414-020-02483-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-020-02483-x